Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap
- PMID: 29439242
- PMCID: PMC5898828
- DOI: 10.1126/science.aad6469
Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap
Abstract
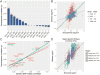
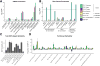
The predisposition to neuropsychiatric disease involves a complex, polygenic, and pleiotropic genetic architecture. However, little is known about how genetic variants impart brain dysfunction or pathology. We used transcriptomic profiling as a quantitative readout of molecular brain-based phenotypes across five major psychiatric disorders-autism, schizophrenia, bipolar disorder, depression, and alcoholism-compared with matched controls. We identified patterns of shared and distinct gene-expression perturbations across these conditions. The degree of sharing of transcriptional dysregulation is related to polygenic (single-nucleotide polymorphism-based) overlap across disorders, suggesting a substantial causal genetic component. This comprehensive systems-level view of the neurobiological architecture of major neuropsychiatric illness demonstrates pathways of molecular convergence and specificity.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures


References
Publication types
MeSH terms
Grants and funding
- R37 MH057881/MH/NIMH NIH HHS/United States
- P50 MH106438/MH/NIMH NIH HHS/United States
- R21 MH103877/MH/NIMH NIH HHS/United States
- U01 MH103365/MH/NIMH NIH HHS/United States
- P50 MH084051/MH/NIMH NIH HHS/United States
- R01 MH110927/MH/NIMH NIH HHS/United States
- F31 AG051381/AG/NIA NIH HHS/United States
- U01 MH103392/MH/NIMH NIH HHS/United States
- U01 MH103346/MH/NIMH NIH HHS/United States
- R01 MH105472/MH/NIMH NIH HHS/United States
- U01 MH103340/MH/NIMH NIH HHS/United States
- P50 MH084053/MH/NIMH NIH HHS/United States
- R01 MH110920/MH/NIMH NIH HHS/United States
- R21 MH105881/MH/NIMH NIH HHS/United States
- P50 MH066392/MH/NIMH NIH HHS/United States
- R03 MH066923/MH/NIMH NIH HHS/United States
- R01 MH085542/MH/NIMH NIH HHS/United States
- P50 MH106934/MH/NIMH NIH HHS/United States
- P50 MH096891/MH/NIMH NIH HHS/United States
- R01 MH094714/MH/NIMH NIH HHS/United States
- R01 MH075916/MH/NIMH NIH HHS/United States
- R01 MH100027/MH/NIMH NIH HHS/United States
- U01 MH103339/MH/NIMH NIH HHS/United States
- R01 MH097276/MH/NIMH NIH HHS/United States
- R03 AG022193/AG/NIA NIH HHS/United States
- P01 AG002219/AG/NIA NIH HHS/United States
- R01 MH093725/MH/NIMH NIH HHS/United States
- P50 AG005138/AG/NIA NIH HHS/United States
- P50 MH096890/MH/NIMH NIH HHS/United States
- R01 ES024988/ES/NIEHS NIH HHS/United States
- R21 MH102791/MH/NIMH NIH HHS/United States
- R01 MH105898/MH/NIMH NIH HHS/United States
- R01 MH109912/MH/NIMH NIH HHS/United States
- R01 MH080405/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

