Polyporales Brown Rot Species Fomitopsis pinicola: Enzyme Activity Profiles, Oxalic Acid Production, and Fe3+-Reducing Metabolite Secretion
- PMID: 29439983
- PMCID: PMC5881074
- DOI: 10.1128/AEM.02662-17
Polyporales Brown Rot Species Fomitopsis pinicola: Enzyme Activity Profiles, Oxalic Acid Production, and Fe3+-Reducing Metabolite Secretion
Abstract
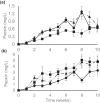
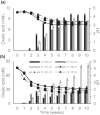
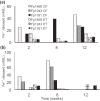
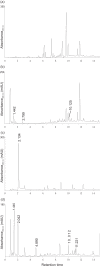
Basidiomycota fungi in the order Polyporales are specified to decomposition of dead wood and woody debris and thereby are crucial players in the degradation of organic matter and cycling of carbon in the forest ecosystems. Polyporales wood-decaying species comprise both white rot and brown rot fungi, based on their mode of wood decay. While the white rot fungi are able to attack and decompose all the lignocellulose biopolymers, the brown rot species mainly cause the destruction of wood polysaccharides, with minor modification of the lignin units. The biochemical mechanism of brown rot decay of wood is still unclear and has been proposed to include a combination of nonenzymatic oxidation reactions and carbohydrate-active enzymes. Therefore, a linking approach is needed to dissect the fungal brown rot processes. We studied the brown rot Polyporales species Fomitopsis pinicola by following mycelial growth and enzyme activity patterns and generating metabolites together with Fenton-promoting Fe3+-reducing activity for 3 months in submerged cultures supplemented with spruce wood. Enzyme activities to degrade hemicellulose, cellulose, proteins, and chitin were produced by three Finnish isolates of F. pinicola Substantial secretion of oxalic acid and a decrease in pH were notable. Aromatic compounds and metabolites were observed to accumulate in the fungal cultures, with some metabolites having Fe3+-reducing activity. Thus, F. pinicola demonstrates a pattern of strong mycelial growth leading to the active production of carbohydrate- and protein-active enzymes, together with the promotion of Fenton biochemistry. Our findings point to fungal species-level "fine-tuning" and variations in the biochemical reactions leading to the brown rot type of wood decay.IMPORTANCEFomitopsis pinicola is a common fungal species in boreal and temperate forests in the Northern Hemisphere encountered as a wood-colonizing saprotroph and tree pathogen, causing a severe brown rot type of wood degradation. However, its lignocellulose-decomposing mechanisms have remained undiscovered. Our approach was to explore both the enzymatic activities and nonenzymatic Fenton reaction-promoting activities (Fe3+ reduction and metabolite production) by cultivating three isolates of F. pinicola in wood-supplemented cultures. Our findings on the simultaneous production of versatile enzyme activities, including those of endoglucanase, xylanase, β-glucosidase, chitinase, and acid peptidase, together with generation of low pH, accumulation of oxalic acid, and Fe3+-reducing metabolites, increase the variations of fungal brown rot decay mechanisms. Furthermore, these findings will aid us in revealing the wood decay proteomic, transcriptomic, and metabolic activities of this ecologically important forest fungal species.
Keywords: Agaricomycetes; Fenton reaction; Polyporales; basidiomycetes; biodegradation; brown rot; fungal enzymes; lignocellulose; oxalic acid; wood decay fungi.
Copyright © 2018 American Society for Microbiology.
Figures



References
-
- Eriksson KEL, Blanchette RA, Ander P. 1990. Microbial and enzymatic degradation of wood and wood components. Springer, Berlin, Germany.
-
- Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A, Asiegbu FO, Baker SE, Barry K, Bendiksby M, Blumentritt M, Coutinho PM, Cullen D, de Vries RP, Gathman A, Goodell B, Henrissat B, Ihrmark K, Kauserud H, Kohler A, LaButti K, Lapidus A, Lavin JL, Lee YH, Lindquist E, Lilly W, Lucas S, Morin E, Murat C, Oguiza JA, Park J, Pisabarro AG, Riley R, Rosling A, Salamov A, Schmidt O, Schmutz J, Skrede I, Stenlid J, Wiebenga A, Xie XF, Kües U, Hibbett DS, Hoffmeister D, Hogberg N, Martin F, Grigoriev IV, Watkinson SC. 2011. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 333:762–765. doi:10.1126/science.1205411. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

