Genomic Predictions and Genome-Wide Association Study of Resistance Against Piscirickettsia salmonis in Coho Salmon (Oncorhynchus kisutch) Using ddRAD Sequencing
- PMID: 29440129
- PMCID: PMC5873909
- DOI: 10.1534/g3.118.200053
Genomic Predictions and Genome-Wide Association Study of Resistance Against Piscirickettsia salmonis in Coho Salmon (Oncorhynchus kisutch) Using ddRAD Sequencing
Abstract
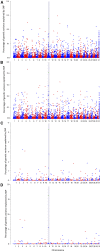
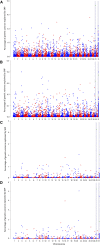
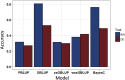
Piscirickettsia salmonis is one of the main infectious diseases affecting coho salmon (Oncorhynchus kisutch) farming, and current treatments have been ineffective for the control of this disease. Genetic improvement for P. salmonis resistance has been proposed as a feasible alternative for the control of this infectious disease in farmed fish. Genotyping by sequencing (GBS) strategies allow genotyping of hundreds of individuals with thousands of single nucleotide polymorphisms (SNPs), which can be used to perform genome wide association studies (GWAS) and predict genetic values using genome-wide information. We used double-digest restriction-site associated DNA (ddRAD) sequencing to dissect the genetic architecture of resistance against P. salmonis in a farmed coho salmon population and to identify molecular markers associated with the trait. We also evaluated genomic selection (GS) models in order to determine the potential to accelerate the genetic improvement of this trait by means of using genome-wide molecular information. A total of 764 individuals from 33 full-sib families (17 highly resistant and 16 highly susceptible) were experimentally challenged against P. salmonis and their genotypes were assayed using ddRAD sequencing. A total of 9,389 SNPs markers were identified in the population. These markers were used to test genomic selection models and compare different GWAS methodologies for resistance measured as day of death (DD) and binary survival (BIN). Genomic selection models showed higher accuracies than the traditional pedigree-based best linear unbiased prediction (PBLUP) method, for both DD and BIN. The models showed an improvement of up to 95% and 155% respectively over PBLUP. One SNP related with B-cell development was identified as a potential functional candidate associated with resistance to P. salmonis defined as DD.
Keywords: GWAS; GenPred; Genomic Selection; Oncorhynchus kisutch; Shared Data Resources; disease resistance; genotyping by sequencing; selective breeding.
Copyright © 2018 Barria et al.
Figures

References
-
- Brieuc M. S. O., Waters C. D., Seeb J. E., and K. A. Naish, 2014. A dense linkage map for Chinook salmon (Oncorhynchus tshawytscha) reveals variable chromosomal divergence after an ancestral whole genome duplication event. G3 Genes Genomes Genet. 4: 447–460. https://doi.org/10.1534/g3.113.009316 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
