Three-step transcriptional priming that drives the commitment of multipotent progenitors toward B cells
- PMID: 29440259
- PMCID: PMC5830925
- DOI: 10.1101/gad.309575.117
Three-step transcriptional priming that drives the commitment of multipotent progenitors toward B cells
Abstract
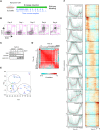
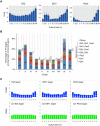
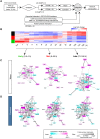
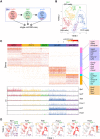
Stem cell fate is orchestrated by core transcription factors (TFs) and epigenetic modifications. Although regulatory genes that control cell type specification are identified, the transcriptional circuit and the cross-talk among regulatory factors during cell fate decisions remain poorly understood. To identify the "time-lapse" TF networks during B-lineage commitment, we used multipotent progenitors harboring a tamoxifen-inducible form of Id3, an in vitro system in which virtually all cells became B cells within 6 d by simply withdrawing 4-hydroxytamoxifen (4-OHT). Transcriptome and epigenome analysis at multiple time points revealed that ∼10%-30% of differentially expressed genes were virtually controlled by the core TFs, including E2A, EBF1, and PAX5. Strikingly, we found unexpected transcriptional priming before the onset of the key TF program. Inhibition of the immediate early genes such as Nr4a2, Klf4, and Egr1 severely impaired the generation of B cells. Integration of multiple data sets, including transcriptome, protein interactome, and epigenome profiles, identified three representative transcriptional circuits. Single-cell RNA sequencing (RNA-seq) analysis of lymphoid progenitors in bone marrow strongly supported the three-step TF network model during specification of multipotent progenitors toward B-cell lineage in vivo. Thus, our findings will provide a blueprint for studying the normal and neoplastic development of B lymphocytes.
Keywords: B-cell differentiation; epigenetics; lineage commitment; single-cell RNA-seq; transcription factor; transcriptional network.
© 2018 Miyai et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


Comment in
-
'Big bang' of B-cell development revealed.Genes Dev. 2018 Jan 15;32(2):93-95. doi: 10.1101/gad.311357.118. Genes Dev. 2018. PMID: 29449365 Free PMC article.
References
-
- Bannister AJ, Kouzarides T. 2005. Reversing histone methylation. Nature 436: 1103–1106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
