Dynamic EBF1 occupancy directs sequential epigenetic and transcriptional events in B-cell programming
- PMID: 29440261
- PMCID: PMC5830932
- DOI: 10.1101/gad.309583.117
Dynamic EBF1 occupancy directs sequential epigenetic and transcriptional events in B-cell programming
Abstract
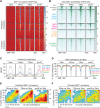
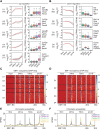
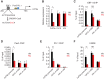
B-cell fate determination requires the action of transcription factors that operate in a regulatory network to activate B-lineage genes and repress lineage-inappropriate genes. However, the dynamics and hierarchy of events in B-cell programming remain obscure. To uncouple the dynamics of transcription factor expression from functional consequences, we generated induction systems in developmentally arrested Ebf1-/- pre-pro-B cells to allow precise experimental control of EBF1 expression in the genomic context of progenitor cells. Consistent with the described role of EBF1 as a pioneer transcription factor, we show in a time-resolved analysis that EBF1 occupancy coincides with EBF1 expression and precedes the formation of chromatin accessibility. We observed dynamic patterns of EBF1 target gene expression and sequential up-regulation of transcription factors that expand the regulatory network at the pro-B-cell stage. A continuous EBF1 function was found to be required for Cd79a promoter activity and for the maintenance of an accessible chromatin domain that is permissive for binding of other transcription factors. Notably, transient EBF1 occupancy was detected at lineage-inappropriate genes prior to their silencing in pro-B cells. Thus, persistent and transient functions of EBF1 allow for an ordered sequence of epigenetic and transcriptional events in B-cell programming.
Keywords: B-cell programming; DNA methylation; EBF1; IRF4; Pax5; chromatin.
© 2018 Li et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Comment in
-
'Big bang' of B-cell development revealed.Genes Dev. 2018 Jan 15;32(2):93-95. doi: 10.1101/gad.311357.118. Genes Dev. 2018. PMID: 29449365 Free PMC article.
References
-
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. 2005. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 121: 295–306. - PubMed
-
- Bain G, Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, et al. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79: 885–892. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
