ATP synthase from Trypanosoma brucei has an elaborated canonical F1-domain and conventional catalytic sites
- PMID: 29440423
- PMCID: PMC5834723
- DOI: 10.1073/pnas.1720940115
ATP synthase from Trypanosoma brucei has an elaborated canonical F1-domain and conventional catalytic sites
Abstract
The structures and functions of the components of ATP synthases, especially those subunits involved directly in the catalytic formation of ATP, are widely conserved in metazoans, fungi, eubacteria, and plant chloroplasts. On the basis of a map at 32.5-Å resolution determined in situ in the mitochondria of Trypanosoma brucei by electron cryotomography, it has been proposed that the ATP synthase in this species has a noncanonical structure and different catalytic sites in which the catalytically essential arginine finger is provided not by the α-subunit adjacent to the catalytic nucleotide-binding site as in all species investigated to date, but rather by a protein, p18, found only in the euglenozoa. A crystal structure at 3.2-Å resolution of the catalytic domain of the same enzyme demonstrates that this proposal is incorrect. In many respects, the structure is similar to the structures of F1-ATPases determined previously. The α3β3-spherical portion of the catalytic domain in which the three catalytic sites are found, plus the central stalk, are highly conserved, and the arginine finger is provided conventionally by the α-subunits adjacent to each of the three catalytic sites found in the β-subunits. Thus, the enzyme has a conventional catalytic mechanism. The structure differs from previous described structures by the presence of a p18 subunit, identified only in the euglenozoa, associated with the external surface of each of the three α-subunits, thereby elaborating the F1-domain. Subunit p18 is a pentatricopeptide repeat (PPR) protein with three PPRs and appears to have no function in the catalytic mechanism of the enzyme.
Keywords: ATP synthase; Trypanosoma brucei; catalytic domain; p18 subunit; structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


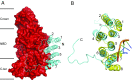
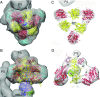
Comment in
-
Structure of the catalytic F1 head of the F1-Fo ATP synthase from Trypanosoma brucei.Proc Natl Acad Sci U S A. 2018 Mar 27;115(13):E2906-E2907. doi: 10.1073/pnas.1801103115. Epub 2018 Mar 9. Proc Natl Acad Sci U S A. 2018. PMID: 29523707 Free PMC article. No abstract available.
Similar articles
-
The F1 -ATPase from Trypanosoma brucei is elaborated by three copies of an additional p18-subunit.FEBS J. 2018 Feb;285(3):614-628. doi: 10.1111/febs.14364. Epub 2017 Dec 30. FEBS J. 2018. PMID: 29247468
-
In situ structure of trypanosomal ATP synthase dimer reveals a unique arrangement of catalytic subunits.Proc Natl Acad Sci U S A. 2017 Jan 31;114(5):992-997. doi: 10.1073/pnas.1612386114. Epub 2017 Jan 17. Proc Natl Acad Sci U S A. 2017. PMID: 28096380 Free PMC article.
-
Characterization of a highly diverged mitochondrial ATP synthase Fo subunit in Trypanosoma brucei.J Biol Chem. 2022 Apr;298(4):101829. doi: 10.1016/j.jbc.2022.101829. Epub 2022 Mar 12. J Biol Chem. 2022. PMID: 35293314 Free PMC article.
-
Redesigned and reversed: architectural and functional oddities of the trypanosomal ATP synthase.Parasitology. 2021 Sep;148(10):1151-1160. doi: 10.1017/S0031182021000202. Epub 2021 Feb 8. Parasitology. 2021. PMID: 33551002 Free PMC article. Review.
-
Structures and interactions of proteins involved in the coupling function of the protonmotive F(o)F(1)-ATP synthase.Curr Protein Pept Sci. 2002 Aug;3(4):451-60. doi: 10.2174/1389203023380558. Curr Protein Pept Sci. 2002. PMID: 12370007 Review.
Cited by
-
The persistent homology of mitochondrial ATP synthases.iScience. 2023 Apr 19;26(5):106700. doi: 10.1016/j.isci.2023.106700. eCollection 2023 May 19. iScience. 2023. PMID: 37250340 Free PMC article.
-
Supramolecular associations between atypical oxidative phosphorylation complexes of Euglena gracilis.J Bioenerg Biomembr. 2021 Jun;53(3):351-363. doi: 10.1007/s10863-021-09882-8. Epub 2021 Mar 1. J Bioenerg Biomembr. 2021. PMID: 33646522 Free PMC article.
-
The atypical subunit composition of respiratory complexes I and IV is associated with original extra structural domains in Euglena gracilis.Sci Rep. 2018 Jun 26;8(1):9698. doi: 10.1038/s41598-018-28039-z. Sci Rep. 2018. PMID: 29946152 Free PMC article.
-
Bioenergetic consequences of FoF1-ATP synthase/ATPase deficiency in two life cycle stages of Trypanosoma brucei.J Biol Chem. 2021 Jan-Jun;296:100357. doi: 10.1016/j.jbc.2021.100357. Epub 2021 Feb 2. J Biol Chem. 2021. PMID: 33539923 Free PMC article.
-
The Mitochondrial Calcium Uniporter Interacts with Subunit c of the ATP Synthase of Trypanosomes and Humans.mBio. 2020 Mar 17;11(2):e00268-20. doi: 10.1128/mBio.00268-20. mBio. 2020. PMID: 32184243 Free PMC article.
References
-
- Walker JE. The ATP synthase: The understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41:1–16. - PubMed
-
- Walker JE. Structure, mechanism, and regulation of ATP synthases. In: Wikström M, editor. Mechanisms of Primary Energy Transduction in Biology. Royal Society of Chemistry; London: 2017. pp. 338–373.
-
- Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8-Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

