Windowed Granger causal inference strategy improves discovery of gene regulatory networks
- PMID: 29440433
- PMCID: PMC5834671
- DOI: 10.1073/pnas.1710936115
Windowed Granger causal inference strategy improves discovery of gene regulatory networks
Abstract
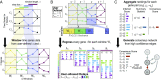
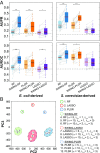
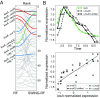
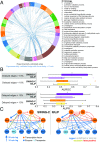
Accurate inference of regulatory networks from experimental data facilitates the rapid characterization and understanding of biological systems. High-throughput technologies can provide a wealth of time-series data to better interrogate the complex regulatory dynamics inherent to organisms, but many network inference strategies do not effectively use temporal information. We address this limitation by introducing Sliding Window Inference for Network Generation (SWING), a generalized framework that incorporates multivariate Granger causality to infer network structure from time-series data. SWING moves beyond existing Granger methods by generating windowed models that simultaneously evaluate multiple upstream regulators at several potential time delays. We demonstrate that SWING elucidates network structure with greater accuracy in both in silico and experimentally validated in vitro systems. We estimate the apparent time delays present in each system and demonstrate that SWING infers time-delayed, gene-gene interactions that are distinct from baseline methods. By providing a temporal framework to infer the underlying directed network topology, SWING generates testable hypotheses for gene-gene influences.
Keywords: Granger causality; gene regulatory networks; machine learning; network inference; time-series analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Reverse engineering gene regulatory networks: coupling an optimization algorithm with a parameter identification technique.BMC Bioinformatics. 2014;15 Suppl 15(Suppl 15):S8. doi: 10.1186/1471-2105-15-S15-S8. Epub 2014 Dec 3. BMC Bioinformatics. 2014. PMID: 25474560 Free PMC article.
-
A novel procedure for statistical inference and verification of gene regulatory subnetwork.BMC Bioinformatics. 2015;16 Suppl 7(Suppl 7):S7. doi: 10.1186/1471-2105-16-S7-S7. Epub 2015 Apr 23. BMC Bioinformatics. 2015. PMID: 25952938 Free PMC article.
-
CaSPIAN: a causal compressive sensing algorithm for discovering directed interactions in gene networks.PLoS One. 2014 Mar 12;9(3):e90781. doi: 10.1371/journal.pone.0090781. eCollection 2014. PLoS One. 2014. PMID: 24622336 Free PMC article.
-
Biological Network Inference and analysis using SEBINI and CABIN.Methods Mol Biol. 2009;541:551-76. doi: 10.1007/978-1-59745-243-4_24. Methods Mol Biol. 2009. PMID: 19381531 Review.
-
Biological network inference for drug discovery.Drug Discov Today. 2013 Mar;18(5-6):256-64. doi: 10.1016/j.drudis.2012.11.001. Epub 2012 Nov 10. Drug Discov Today. 2013. PMID: 23147668 Review.
Cited by
-
Causal network inference from gene transcriptional time-series response to glucocorticoids.PLoS Comput Biol. 2021 Jan 29;17(1):e1008223. doi: 10.1371/journal.pcbi.1008223. eCollection 2021 Jan. PLoS Comput Biol. 2021. PMID: 33513136 Free PMC article.
-
Network inference with Granger causality ensembles on single-cell transcriptomics.Cell Rep. 2022 Feb 8;38(6):110333. doi: 10.1016/j.celrep.2022.110333. Cell Rep. 2022. PMID: 35139376 Free PMC article.
-
Murine single-cell RNA-seq reveals cell-identity- and tissue-specific trajectories of aging.Genome Res. 2019 Dec;29(12):2088-2103. doi: 10.1101/gr.253880.119. Epub 2019 Nov 21. Genome Res. 2019. PMID: 31754020 Free PMC article.
-
Hybrid analysis of gene dynamics predicts context-specific expression and offers regulatory insights.Bioinformatics. 2019 Nov 1;35(22):4671-4678. doi: 10.1093/bioinformatics/btz256. Bioinformatics. 2019. PMID: 30994899 Free PMC article.
-
From time-series transcriptomics to gene regulatory networks: A review on inference methods.PLoS Comput Biol. 2023 Aug 10;19(8):e1011254. doi: 10.1371/journal.pcbi.1011254. eCollection 2023 Aug. PLoS Comput Biol. 2023. PMID: 37561790 Free PMC article. Review.
References
-
- Nagoshi E, et al. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. - PubMed
-
- Jiang YJ, et al. Notch signalling and the synchronization of the somite segmentation clock. Nature. 2000;408:475–479. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

