The Birth and Demise of the IS Apl1- mcr-1-IS Apl1 Composite Transposon: the Vehicle for Transferable Colistin Resistance
- PMID: 29440577
- PMCID: PMC5821093
- DOI: 10.1128/mBio.02381-17
The Birth and Demise of the IS Apl1- mcr-1-IS Apl1 Composite Transposon: the Vehicle for Transferable Colistin Resistance
Abstract
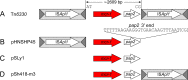
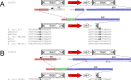
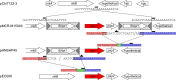
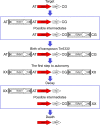
The origin and mobilization of the ~2,609-bp DNA segment containing the mobile colistin resistance gene mcr-1 continue to be sources of uncertainty, but recent evidence suggests that the gene originated in Moraxella species. Moreover mcr-1 can be mobilized as an ISApl1-flanked composite transposon (Tn6330), but many sequences have been identified without ISApl1 or with just a single copy (single ended). To further clarify the origins and mobilization of mcr-1, we employed the Geneious R8 software suite to comprehensively analyze the genetic environment of every complete mcr-1 structure deposited in GenBank as of this writing (September 2017) both with and without associated ISApl1 (n = 273). This revealed that the 2,609-bp mcr-1 structure was likely mobilized from a close relative of a novel species of Moraxella containing a chromosomal region sharing >96% nucleotide identity with the canonical sequence. This chromosomal region is bounded by AT and CG dinucleotides, which have been described on the inside ends (IE) of all intact Tn6330 described to date and represent the ancestral 2-bp target site duplications (TSDs) generated by ISApl1 transposition. We further demonstrate that all mcr-1 structures with just one ISApl1 copy or with no ISApl1 copies were formed by deletion of ISApl1 from the ancestral Tn6330, likely by a process related to the "copy-out-paste-in" transposition mechanism. Finally, we show that only the rare examples of single-ended structures that have retained a portion of the excised downstream ISApl1 including the entire inverted right repeat might be capable of mobilization.IMPORTANCE A comprehensive analysis of all intact mcr-1 sequences in GenBank was used to identify a region on the chromosome of a novel Moraxella species with remarkable homology to the canonical mcr-1 structure and that likely represents the origin of this important gene. These data also demonstrate that all mcr-1 structures lacking one or both flanking ISApl1 were formed from ancestral composite transposons that subsequently lost the insertion sequences by a process of abortive transposition. This observation conclusively shows that mobilization of mcr-1 occurs as part of a composite transposon and that structures lacking the downstream ISApl1 are not capable of mobilization.
Keywords: colistin resistance; composite transposon formation; drug resistance evolution; insertion sequence; transposon decay.
Copyright © 2018 Snesrud et al.
Figures





References
-
- Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M. 2014. Everyman’s guide to bacterial insertion sequences, p 555–590. In Craig NL, Chandler M, Gellert M, Lambowitz AM, Rice PA, Sandmeyer SB (ed), Mobile DNA III, 3rd ed. ASM Press, Washington DC.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

