Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells
- PMID: 29440634
- PMCID: PMC5811451
- DOI: 10.1038/s41467-018-02993-8
Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells
Abstract
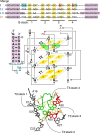
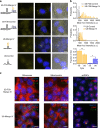
Despite having many key roles in cellular biology, directly imaging biologically important RNAs has been hindered by a lack of fluorescent tools equivalent to the fluorescent proteins available to study cellular proteins. Ideal RNA labelling systems must preserve biological function, have photophysical properties similar to existing fluorescent proteins, and be compatible with established live and fixed cell protein labelling strategies. Here, we report a microfluidics-based selection of three new high-affinity RNA Mango fluorogenic aptamers. Two of these are as bright or brighter than enhanced GFP when bound to TO1-Biotin. Furthermore, we show that the new Mangos can accurately image the subcellular localization of three small non-coding RNAs (5S, U6, and a box C/D scaRNA) in fixed and live mammalian cells. These new aptamers have many potential applications to study RNA function and dynamics both in vitro and in mammalian cells.
Conflict of interest statement
P.J.U., M.R., S.J., A.A., A.Ab. and S.S.S.P. have filed a provisional patent on aspects of this work. The remaining authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

