The effects of death and post-mortem cold ischemia on human tissue transcriptomes
- PMID: 29440659
- PMCID: PMC5811508
- DOI: 10.1038/s41467-017-02772-x
The effects of death and post-mortem cold ischemia on human tissue transcriptomes
Abstract
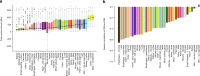
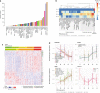
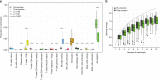
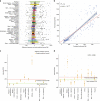
Post-mortem tissues samples are a key resource for investigating patterns of gene expression. However, the processes triggered by death and the post-mortem interval (PMI) can significantly alter physiologically normal RNA levels. We investigate the impact of PMI on gene expression using data from multiple tissues of post-mortem donors obtained from the GTEx project. We find that many genes change expression over relatively short PMIs in a tissue-specific manner, but this potentially confounding effect in a biological analysis can be minimized by taking into account appropriate covariates. By comparing ante- and post-mortem blood samples, we identify the cascade of transcriptional events triggered by death of the organism. These events do not appear to simply reflect stochastic variation resulting from mRNA degradation, but active and ongoing regulation of transcription. Finally, we develop a model to predict the time since death from the analysis of the transcriptome of a few readily accessible tissues.
Conflict of interest statement
P.G.F., C.O., and P.O. are partners of Bioinf2Bio. The remaining authors declare no competing financial interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

