Folic Acid Exposure Rescues Spina Bifida Aperta Phenotypes in Human Induced Pluripotent Stem Cell Model
- PMID: 29440666
- PMCID: PMC5811493
- DOI: 10.1038/s41598-018-21103-8
Folic Acid Exposure Rescues Spina Bifida Aperta Phenotypes in Human Induced Pluripotent Stem Cell Model
Abstract
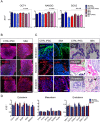
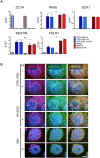
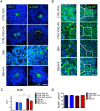
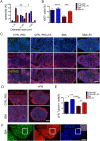
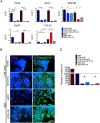
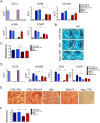
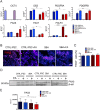
Neural tube defects (NTDs) are severe congenital abnormalities, caused by failed closure of neural tube during early embryonic development. Periconceptional folic acid (FA) supplementation greatly reduces the risk of NTDs. However, the molecular mechanisms behind NTDs and the preventive role of FA remain unclear. Here, we use human induced pluripotent stem cells (iPSCs) derived from fetuses with spina bifida aperta (SBA) to study the pathophysiology of NTDs and explore the effects of FA exposure. We report that FA exposure in SBA model is necessary for the proper formation and maturation of neural tube structures and robust differentiation of mesodermal derivatives. Additionally, we show that the folate antagonist methotrexate dramatically affects the formation of neural tube structures and FA partially reverts this aberrant phenotype. In conclusion, we present a novel model for human NTDs and provide evidence that it is a powerful tool to investigate the molecular mechanisms underlying NTDs, test drugs for therapeutic approaches.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Altered expression of 14-3-3ζ protein in spinal cords of rat fetuses with spina bifida aperta.PLoS One. 2013 Aug 6;8(8):e70457. doi: 10.1371/journal.pone.0070457. Print 2013. PLoS One. 2013. PMID: 23936434 Free PMC article.
-
Cellular mechanisms underlying Pax3-related neural tube defects and their prevention by folic acid.Dis Model Mech. 2019 Nov 22;12(11):dmm042234. doi: 10.1242/dmm.042234. Dis Model Mech. 2019. PMID: 31636139 Free PMC article.
-
Fetal neural tube stem cells from Pax3 mutant mice proliferate, differentiate, and form synaptic connections when stimulated with folic acid.Stem Cells Dev. 2012 Jan 20;21(2):321-30. doi: 10.1089/scd.2011.0100. Epub 2011 Jun 9. Stem Cells Dev. 2012. PMID: 21521032
-
Spina bifida and other neural tube defects.Curr Probl Pediatr. 2000 Nov-Dec;30(10):313-32. doi: 10.1067/mpp.2000.112052. Curr Probl Pediatr. 2000. PMID: 11147289 Review.
-
Mini-review: toward understanding mechanisms of genetic neural tube defects in mice.Teratology. 1999 Nov;60(5):292-305. doi: 10.1002/(SICI)1096-9926(199911)60:5<292::AID-TERA10>3.0.CO;2-6. Teratology. 1999. PMID: 10525207 Review.
Cited by
-
Distinct effects of interleukin-6 and interferon-γ on differentiating human cortical neurons.Brain Behav Immun. 2022 Jul;103:97-108. doi: 10.1016/j.bbi.2022.04.007. Epub 2022 Apr 13. Brain Behav Immun. 2022. PMID: 35429607 Free PMC article.
-
Up-regulation of RNA Binding Proteins Contributes to Folate Deficiency-Induced Neural Crest Cells Dysfunction.Int J Biol Sci. 2020 Jan 1;16(1):85-98. doi: 10.7150/ijbs.33976. eCollection 2020. Int J Biol Sci. 2020. PMID: 31892848 Free PMC article.
-
A Shared Pathogenic Mechanism for Valproic Acid and SHROOM3 Knockout in a Brain Organoid Model of Neural Tube Defects.Cells. 2023 Jun 23;12(13):1697. doi: 10.3390/cells12131697. Cells. 2023. PMID: 37443734 Free PMC article.
-
Validation of the Fetal Lamb Model of Spina Bifida.Sci Rep. 2019 Jun 27;9(1):9327. doi: 10.1038/s41598-019-45819-3. Sci Rep. 2019. PMID: 31249378 Free PMC article.
-
Engineered tissues and strategies to overcome challenges in drug development.Adv Drug Deliv Rev. 2020;158:116-139. doi: 10.1016/j.addr.2020.09.012. Epub 2020 Sep 26. Adv Drug Deliv Rev. 2020. PMID: 32987094 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

