Autophagy and Alzheimer's Disease: From Molecular Mechanisms to Therapeutic Implications
- PMID: 29441009
- PMCID: PMC5797541
- DOI: 10.3389/fnagi.2018.00004
Autophagy and Alzheimer's Disease: From Molecular Mechanisms to Therapeutic Implications
Abstract
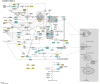
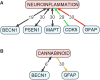
Alzheimer's disease (AD) is the most common cause of progressive dementia in the elderly. It is characterized by a progressive and irreversible loss of cognitive abilities and formation of senile plaques, composed mainly of amyloid β (Aβ), and neurofibrillary tangles (NFTs), composed of tau protein, in the hippocampus and cortex of afflicted humans. In brains of AD patients the metabolism of Aβ is dysregulated, which leads to the accumulation and aggregation of Aβ. Metabolism of Aβ and tau proteins is crucially influenced by autophagy. Autophagy is a lysosome-dependent, homeostatic process, in which organelles and proteins are degraded and recycled into energy. Thus, dysfunction of autophagy is suggested to lead to the accretion of noxious proteins in the AD brain. In the present review, we describe the process of autophagy and its importance in AD. Additionally, we discuss mechanisms and genes linking autophagy and AD, i.e., the mTOR pathway, neuroinflammation, endocannabinoid system, ATG7, BCL2, BECN1, CDK5, CLU, CTSD, FOXO1, GFAP, ITPR1, MAPT, PSEN1, SNCA, UBQLN1, and UCHL1. We also present pharmacological agents acting via modulation of autophagy that may show promise in AD therapy. This review updates our knowledge on autophagy mechanisms proposing novel therapeutic targets for the treatment of AD.
Keywords: Alzheimer’s disease; amyloid beta; autophagy; tau.
Figures
References
-
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. (2002). Transport from the Trans Golgi Network to Lysosomes. New York, NY: Garland Science.
-
- Armstrong R. A. (2009). The molecular biology of senile plaques and neurofibrillary tangles in Alzheimer’s disease. Folia Neuropathol. 47 289–299. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

