Parallel lives of symbionts and hosts: chemical mutualism in marine animals
- PMID: 29441375
- PMCID: PMC6025756
- DOI: 10.1039/c7np00053g
Parallel lives of symbionts and hosts: chemical mutualism in marine animals
Abstract
Covering: up to 2018 Symbiotic microbes interact with animals, often by producing natural products (specialized metabolites; secondary metabolites) that exert a biological role. A major goal is to determine which microbes produce biologically important compounds, a deceptively challenging task that often rests on correlative results, rather than hypothesis testing. Here, we examine the challenges and successes from the perspective of marine animal-bacterial mutualisms. These animals have historically provided a useful model because of their technical accessibility. By comparing biological systems, we suggest a common framework for establishing chemical interactions between animals and microbes.
Figures
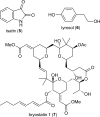

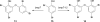




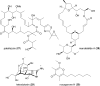

References
-
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Science. 2005;307:1915–1920. - PubMed
-
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, Fitzgerald MG, Fulton RS, Giglio MG, Hallsworth-Pepin K, Lobos EA, Madupu R, Magrini V, Martin JC, Mitreva M, Muzny DM, Sodergren EJ, Versalovic J, Wollam AM, Worley KC, Wortman JR, Young SK, Zeng Q, Aagaard KM, Abolude OO, Allen-Vercoe E, Alm EJ, Alvarado L, Andersen GL, Anderson S, Appelbaum E, Arachchi HM, Armitage G, Arze CA, Ayvaz T, Baker CC, Begg L, Belachew T, Bhonagiri V, Bihan M, Blaser MJ, Bloom T, Bonazzi V, Paul Brooks J, Buck GA, Buhay CJ, Busam DA, Campbell JL, Canon SR, Cantarel BL, Chain PSG, Chen IMA, Chen L, Chhibba S, Chu K, Ciulla DM, Clemente JC, Clifton SW, Conlan S, Crabtree J, Cutting MA, Davidovics NJ, Davis CC, Desantis TZ, Deal C, Delehaunty KD, Dewhirst FE, Deych E, Ding Y, Dooling DJ, Dugan SP, Michael Dunne W, Scott Durkin A, Edgar RC, Erlich RL, Farmer CN, Farrell RM, Faust K, Feldgarden M, Felix VM, Fisher S, Fodor AA, Forney LJ, Foster L, Di Francesco V, Friedman J, Friedrich DC, Fronick CC, Fulton LL, Gao H, Garcia N, Giannoukos G, Giblin C, Giovanni MY, Goldberg JM, Goll J, Gonzalez A, Griggs A, Gujja S, Kinder Haake S, Haas BJ, Hamilton HA, Harris EL, Hepburn TA, Herter B, Hoffmann DE, Holder ME, Howarth C, Huang KH, Huse SM, Izard J, Jansson JK, Jiang H, Jordan C, Joshi V, Katancik JA, Keitel WA, Kelley ST, Kells C, King NB, Knights D, Kong HH, Koren O, Koren S, Kota KC, Kovar CL, Kyrpides NC, La Rosa PS, Lee SL, Lemon KP, Lennon N, Lewis CM, Lewis L, Ley RE, Li K, Liolios K, Liu B, Liu Y, Lo CC, Lozupone CA, Dwayne Lunsford R, Madden T, Mahurkar AA, Mannon PJ, Mardis ER, Markowitz VM, Mavromatis K, McCorrison JM, McDonald D, McEwen J, McGuire AL, McInnes P, Mehta T, Mihindukulasuriya KA, Miller JR, Minx PJ, Newsham I, Nusbaum C, Oglaughlin M, Orvis J, Pagani I, Palaniappan K, Patel SM, Pearson M, Peterson J, Podar M, Pohl C, Pollard KS, Pop M, Priest ME, Proctor LM, Qin X, Raes J, Ravel J, Reid JG, Rho M, Rhodes R, Riehle KP, Rivera MC, Rodriguez-Mueller B, Rogers YH, Ross MC, Russ C, Sanka RK, Sankar P, Fah Sathirapongsasuti J, Schloss JA, Schloss PD, Schmidt TM, Scholz M, Schriml L, Schubert AM, Segata N, Segre JA, Shannon WD, Sharp RR, Sharpton TJ, Shenoy N, Sheth NU, Simone GA, Singh I, Smillie CS, Sobel JD, Sommer DD, Spicer P, Sutton GG, Sykes SM, Tabbaa DG, Thiagarajan M, Tomlinson CM, Torralba M, Treangen TJ, Truty RM, Vishnivetskaya TA, Walker J, Wang L, Wang Z, Ward DV, Warren W, Watson MA, Wellington C, Wetterstrand KA, White JR, Wilczek-Boney K, Wu Y, Wylie KM, Wylie T, Yandava C, Ye L, Ye Y, Yooseph S, Youmans BP, Zhang L, Zhou Y, Zhu Y, Zoloth L, Zucker JD, Birren BW, Gibbs RA, Highlander SK, Methé BA, Nelson KE, Petrosino JF, Weinstock GM, Wilson RK, White O. Nature. 2012;486:207–214.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

