A Comparison of HLA Molecular Mismatch Methods to Determine HLA Immunogenicity
- PMID: 29443827
- PMCID: PMC6072378
- DOI: 10.1097/TP.0000000000002117
A Comparison of HLA Molecular Mismatch Methods to Determine HLA Immunogenicity
Abstract
Background: Antibody-mediated rejection is a major cause of premature graft loss in kidney transplantation. Multiple scoring systems are available to assess the HLA mismatch between donors and recipients at the molecular level; however, their correlation with the development of de novo donor-specific antibody (dnDSA) has not been compared in recipients on active immunosuppression.
Methods: HLA-DRβ1/3/4/5/DQα1β1 molecular mismatch was determined using eplet analysis, amino acid mismatch, and electrostatic mismatch for 596 renal transplant recipients and correlated with HLA-DR/DQ dnDSA development. The molecular mismatch scores were evaluated in multivariate models of posttransplant dnDSA-free survival.
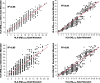
Results: Eplet mismatch correlated with amino acid mismatch and electrostatic mismatch (R = 0.85-0.96). HLA-DR dnDSA-free survival correlated with HLA-DR eplet mismatch (hazards ratio [HR], 2.50 per 10 eplets mismatched; P < 0.0001), amino acid mismatch (HR, 1.49 per 10 amino acids mismatched; P < 0.0001), and electrostatic mismatch (HR, 1.23 per 10 units mismatched; P < 0.0001). HLA-DQ dnDSA-free survival correlated with HLA-DQ eplet mismatch (HR, 1.98 per 10 eplets mismatched; P < 0.0001), amino acid mismatch (HR, 1.24 per 10 amino acids mismatched; P < 0.0001), and electrostatic mismatch (HR, 1.14 per 10 units mismatched; P < 0.0001). All 3 methods were significant multivariate correlates of dnDSA development after adjustment for recipient age, baseline immunosuppression, and nonadherence.
Conclusions: HLA molecular mismatch represents a precise method of alloimmune risk assessment for renal transplant patients. The method used to determine the molecular mismatch is likely to be driven by familiarity and ease of use as highly correlated results are produced by each method.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Wiebe C, Gibson IW, Blydt-Hansen TD, et al. Rates and determinants of progression to graft failure in kidney allograft recipients with de novo donor-specific antibody. Am J Transplant. 2015;15:2921–2930. - PubMed
-
- El-Zoghby ZM, Stegall MD, Lager DJ. Identifying specific causes of kidney allograft loss. Am J Transplant. 2009;9:527–535. - PubMed
-
- Terasaki PI. Humoral theory of transplantation. Am J Transplant. 2003;3:665–673. - PubMed
-
- Wiebe C, Nickerson P. Posttransplant monitoring of de novo human leukocyte antigen donor-specific antibodies in kidney transplantation. Curr Opin Organ Transplant. 2013;18:470–477. - PubMed
-
- Archdeacon P, Chan M, Neuland C, et al. Summary of FDA antibody-mediated rejection workshop. Am J Transplant. 2011;11:896–906. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials