PNLDC1, mouse pre-piRNA Trimmer, is required for meiotic and post-meiotic male germ cell development
- PMID: 29444933
- PMCID: PMC5836094
- DOI: 10.15252/embr.201744957
PNLDC1, mouse pre-piRNA Trimmer, is required for meiotic and post-meiotic male germ cell development
Abstract
PIWI-interacting RNAs (piRNAs) are germ cell-specific small RNAs essential for retrotransposon gene silencing and male germ cell development. In piRNA biogenesis, the endonuclease MitoPLD/Zucchini cleaves long, single-stranded RNAs to generate 5' termini of precursor piRNAs (pre-piRNAs) that are consecutively loaded into PIWI-family proteins. Subsequently, these pre-piRNAs are trimmed at their 3'-end by an exonuclease called Trimmer. Recently, poly(A)-specific ribonuclease-like domain-containing 1 (PNLDC1) was identified as the pre-piRNA Trimmer in silkworms. However, the function of PNLDC1 in other species remains unknown. Here, we generate Pnldc1 mutant mice and analyze small RNAs in their testes. Our results demonstrate that mouse PNLDC1 functions in the trimming of both embryonic and post-natal pre-piRNAs. In addition, piRNA trimming defects in embryonic and post-natal testes cause impaired DNA methylation and reduced MIWI expression, respectively. Phenotypically, both meiotic and post-meiotic arrests are evident in the same individual Pnldc1 mutant mouse. The former and latter phenotypes are similar to those of MILI and MIWI mutant mice, respectively. Thus, PNLDC1-mediated piRNA trimming is indispensable for the function of piRNAs throughout mouse spermatogenesis.
Keywords: PIWI; PNLDC1; Trimmer; piRNA; spermatogenesis.
© 2018 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
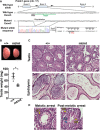
Scheme around exon 3 of mouse Pnldc1 and its targeted locus. PAM and gRNA‐targeted sequences are underlined in black and green, respectively. An 11‐nt deletion in the gRNA‐targeted region and premature stop codon in Pnldc1 mt/mt cDNA in exon 5 were confirmed by sequencing.
Testicular sizes and weights of adult control and Pnldc1 mt/mt mice (mean ± SD, n = 6, *P = 0.0003 by t‐test). Scale bar: 2 mm.
Hematoxylin‐ and eosin‐stained sections of adult control and Pnldc1 mt/mt testes and epididymides. Scale bar: 50 μm.
Representative images of meiotic and post‐meiotic arrest in Pnldc1 mt/mt testes. Scale bar: 50 μm. Spermatocyte (SC), round spermatid (RS), and elongating spermatid (ES) were indicated by yellow and blue arrowheads, respectively.

- A
Body weights of adult control and Pnldc1 mt/mt mice (n = 4).
- B
Scheme around exon 7 in Pnldc1 mice and its targeted locus. PAM and gRNA‐targeted sequences are underlined in black and green, respectively. A retrotransposon sequence was inserted with a 12‐bp deletion at the gRNA‐targeting region (red characters). Genotyping primers are labeled by black arrows.
- C, D
The inserted 836‐bp retrotransposon sequence was confirmed by sequencing (C) and genotyping (D).
- E
Testicular sizes in adult control and Pnldc1 exon 7 mutant mice. Scale bar: 2 mm.
- F
Hematoxylin‐ and eosin‐stained sections of testes and epididymides of adult control and Pnldc1 exon 7 mutant mice. Scale bar: 50 μm.

- A
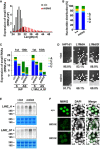
Expression of 24‐ to 50‐nt small RNAs in control and Pnldc1 mt/mt embryonic testes. The small RNAs from E16.5 control and Pnldc1 mt/mt testes were analyzed after removal of rRNA and miRNA mapped reads by piPipes. The 24‐ to 50‐nt small RNA reads were mapped to the mouse genome (mm9). Black and red bars indicate the control and Pnldc1 mt/mt data, respectively.
- B, C
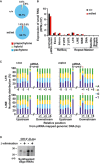
Bisulfite sequencing analysis of H19 DMR, IAP1d1, L1Md_A, and L1Md_Gf genes from purified 10‐day‐old male germ cells from control and Pnldc1 mt/mt testes. DNA methylation of Pnldc1 mt/mt mice (B) and exon 7 mutant mice (C) is shown.
- D
RT‐qPCR analysis of IAP1d1, L1Md_A, and L1Md_Gf transcripts in testes from 14‐day‐old control and Pnldc1 mt/mt mice. Expression levels were normalized to that of β‐actin. Bars show mean ± SEM (n = 4). (P = 0.91 (IAP1d1), **P = 0.004 (L1Md_A), *P = 0.024 (L1Md_Gf) by t‐test).
- E
Western blotting analysis of MIWI2 in E16.5 testes. β‐ACTIN was used as a loading control.
Length distribution of repetitive sequence‐derived small RNAs from E16.5 control and Pnldc1 mt/mt testes. The small RNAs were analyzed after ribosomal RNA (rRNA) and microRNA (miRNA) mapped reads were removed by piPipes. Black and red bars show the control and Pnldc1 mt/mt data, respectively.
Nucleotide distribution of the first nucleotide of repetitive sequence‐derived piRNAs.
Expression and nucleotide distribution of small RNAs corresponding to IAP (M17551) and two LINE‐1 retrotransposon (L1Md_A (M13002) and L1Md_Gf (D84391)) sequences.
Bisulfite sequencing analysis of the IAP1d1, L1Md_A, and L1Md_Gf genes from purified male germ cells in testes from 10‐day‐old control and Pnldc1 mt/mt mice.
Northern blot analysis of L1Md_A and L1Md_Gf transcripts in testes from 14‐day‐old control and Pnldc1 mt/mt mice.
Immunostaining of MIWI2 in E16.5 control and Pnldc1 mt/mt testes. DNA was stained by DAPI. Scale bar: 10 μm.

- A
The RNAs that co‐precipitated with MILI and MIWI2 were purified and separated by 15% denatured acrylamide gel electrophoresis after 32P‐end‐labeling. Immunoprecipitated MILI and MIWI2 proteins were detected by WB with indicated antibodies.
- B–E
Length and nucleotide distributions of MILI‐ and MIWI2‐bound small RNAs from the control and Pnldc1 mt/mt embryonic testes. MILI‐bound small RNAs length distributions (B) and nucleotide distributions (C) are shown by bar graphs. MIWI2‐bound small RNAs length distributions (D) and nucleotide distributions (E) are shown by bar graphs.
Percentages of pachytene and pre‐pachytene piRNAs.
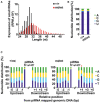
Expression of 24‐ to 50‐nt small RNAs in control and Pnldc1 mt/mt post‐natal testes. Small RNAs from post‐natal day 24 control and Pnldc1 mt/mt testes were analyzed after the removal of rRNA and miRNA mapped reads by piPipes. The 24‐ to 50‐nt small RNA reads were mapped to the mouse genome (mm9). Black and red bars show the control and Pnldc1 mt/mt data, respectively.
Nucleotide distribution around the small RNAs. Small RNA data corresponding to LTR (top) and LINE (bottom) from E16.5 control and Pnldc1 mt/mt testes are shown. Asterisks (*) indicate strong T bias at the +1 position.
32P‐end‐labeled synthesized oligo‐RNAs with or without 2′‐O‐methylation at 3′ end were separated by 12% denatured acrylamide gels with or without β‐elimination treatment.
Length distribution of small RNAs derived from pachytene piRNA clusters from the testes of 24‐day‐old control and Pnldc1 mt/mt mice. The small RNAs were analyzed after rRNA and miRNA mapped reads were removed by piPipes. Black and red bars show the control and Pnldc1 mt/mt data, respectively.
Nucleotide distribution of the first nucleotide of the small RNAs.
Nucleotide distribution around the small RNAs. Asterisk (*) indicates strong T bias at the +1 position.
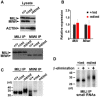
- A, B
Expression analyses of MILI and MIWI in 24‐day‐old testes by Western blotting (A) and RT‐qPCR (B). (A) β‐ACTIN was for the loading control. (B) Relative expression was normalized to that of β‐Actin. Bars show mean ± SEM (n = 3). P = 0.64 for Mili, P = 0.83 for Miwi by t‐test.
- C
MILI‐ and MIWI‐bound small RNAs in testes from 24‐day‐old control and Pnldc1 mt/m mice. The RNAs that co‐precipitated with MILI and MIWI were purified and separated by 15% denatured acrylamide gel electrophoresis after 32P‐end‐labeling.
- D
β‐elimination assay. 32P‐end‐labeled MILI‐bound piRNAs from control and Pnldc1 mt/mt 24‐day‐old testes were separated by 12% denatured acrylamide gels with or without β‐elimination treatment.
Comment in
-
Trimming it short: PNLDC1 is required for piRNA maturation during mouse spermatogenesis.EMBO Rep. 2018 Mar;19(3):e45824. doi: 10.15252/embr.201845824. Epub 2018 Feb 19. EMBO Rep. 2018. PMID: 29459487 Free PMC article.
References
-
- Saito K, Siomi MC (2010) Small RNA‐mediated quiescence of transposable elements in animals. Dev Cell 19: 687–697 - PubMed
-
- Aravin AA, Hannon GJ (2008) Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol 73: 283–290 - PubMed
-
- Iwasaki YW, Siomi MC, Siomi H (2015) PIWI‐interacting RNA: its biogenesis and functions. Annu Rev Biochem 84: 405–433 - PubMed
-
- Siomi MC, Sato K, Pezic D, Aravin AA (2011) PIWI‐interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

