53BP1 can limit sister-chromatid rupture and rearrangements driven by a distinct ultrafine DNA bridging-breakage process
- PMID: 29445165
- PMCID: PMC5813243
- DOI: 10.1038/s41467-018-03098-y
53BP1 can limit sister-chromatid rupture and rearrangements driven by a distinct ultrafine DNA bridging-breakage process
Abstract
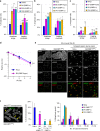
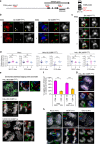
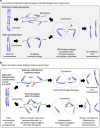
Chromosome missegregation acts as one of the driving forces for chromosome instability and cancer development. Here, we find that in human cancer cells, HeLa and U2OS, depletion of 53BP1 (p53-binding protein 1) exacerbates chromosome non-disjunction resulting from a new type of sister-chromatid intertwinement, which is distinct from FANCD2-associated ultrafine DNA bridges (UFBs) induced by replication stress. Importantly, the sister DNA intertwinements trigger gross chromosomal rearrangements through a distinct process, named sister-chromatid rupture and bridging. In contrast to conventional anaphase bridge-breakage models, we demonstrate that chromatid axes of the intertwined sister-chromatids rupture prior to the breakage of the DNA bridges. Consequently, the ruptured sister arms remain tethered and cause signature chromosome rearrangements, including whole-arm (Robertsonian-like) translocation/deletion and isochromosome formation. Therefore, our study reveals a hitherto unreported chromatid damage phenomenon mediated by sister DNA intertwinements that may help to explain the development of complex karyotypes in tumour cells.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

