The Dynamic Distribution of Small-Tail Han Sheep Microbiota across Different Intestinal Segments
- PMID: 29445360
- PMCID: PMC5797768
- DOI: 10.3389/fmicb.2018.00032
The Dynamic Distribution of Small-Tail Han Sheep Microbiota across Different Intestinal Segments
Abstract
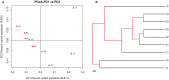
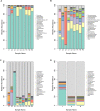
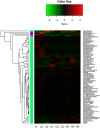
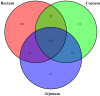
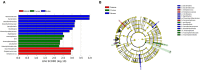
The sheep intestinal tract is characterized by a diverse microbial ecosystem that is vital for the host to digest diet material. The importance of gut microbiota (GM) of animals has also been widely acknowledged because of its pivotal roles in the health and well-being of animals. However, there are no relevant studies on GM of small-tail Han sheep, a superior mutton variety domestic in China. In this study, the structure and distribution of gut microflora were studied by high-throughput sequencing technology. Results showed a significant difference between jejunum and cecum, jejunum, and rectum. Meanwhile, the cecum and rectum not only display higher species richness but also exhibit higher similarity of the bacterial diversity than that of the jejunum based on the results of abundance-based coverage estimator (ACE), Chao1, and Shannon indexes. Firmicutes and Bacteroidetes were the predominant phyla in cecum and rectum, while higher relative abundances of Firmicutes and Cyanobacteria were observed in jejunum. At the genus level, Bacteroidetes, Ruminococcus, Lactobacillus, Flavonifractor, and Clostridium were the dominant genera in the cecum and rectum. An obvious dynamic distribution of Lactobacillus is continuously decreasing from the jejunum to the cecum, then to the rectum, whereas the result of Bacteroides is completely inverse. In addition, this study also found many kinds of bacteria associated with the production of volatile fatty acids (VFA) colonized in the large intestine. This study is the first to investigate the distribution of intestinal flora in small-tail Han sheep. The findings provide an important indication for diagnosis and treatment of intestinal diseases in small-tail Han sheep, as well as offer a direction for the development of intestinal microecological preparations.
Keywords: dynamic distribution; gut microbiota; high-throughput sequencing; small-tail Han sheep; volatile fatty acids.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

