Fe-S Clusters Emerging as Targets of Therapeutic Drugs
- PMID: 29445445
- PMCID: PMC5763138
- DOI: 10.1155/2017/3647657
Fe-S Clusters Emerging as Targets of Therapeutic Drugs
Abstract
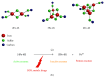
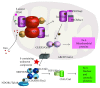
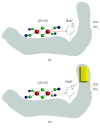
Fe-S centers exhibit strong electronic plasticity, which is of importance for insuring fine redox tuning of protein biological properties. In accordance, Fe-S clusters are also highly sensitive to oxidation and can be very easily altered in vivo by different drugs, either directly or indirectly due to catabolic by-products, such as nitric oxide species (NOS) or reactive oxygen species (ROS). In case of metal ions, Fe-S cluster alteration might be the result of metal liganding to the coordinating sulfur atoms, as suggested for copper. Several drugs presented through this review are either capable of direct interaction with Fe-S clusters or of secondary Fe-S clusters alteration following ROS or NOS production. Reactions leading to Fe-S cluster disruption are also reported. Due to the recent interest and progress in Fe-S biology, it is very likely that an increasing number of drugs already used in clinics will emerge as molecules interfering with Fe-S centers in the near future. Targeting Fe-S centers could also become a promising strategy for drug development.
Figures
References
-
- Golinelli-Cohen M.-P., Bouton C. Fe-S proteins acting as redox switch: new key actors of cellular adaptive responses. Current Chemical Biology. 2017;11:1–19. doi: 10.2174/2212796811666170406163809. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

