Positive and Negative Allosteric Modulators of N-Methyl-d-aspartate (NMDA) Receptors: Structure-Activity Relationships and Mechanisms of Action
- PMID: 29446949
- PMCID: PMC6368479
- DOI: 10.1021/acs.jmedchem.7b01640
Positive and Negative Allosteric Modulators of N-Methyl-d-aspartate (NMDA) Receptors: Structure-Activity Relationships and Mechanisms of Action
Abstract
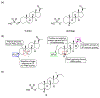
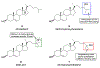
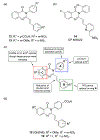
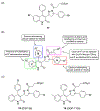
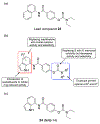
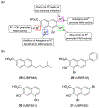
Excitatory activity in the CNS is predominately mediated by l-glutamate through several families of l-glutamate neurotransmitter receptors. Of these, the N-methyl-d-aspartate receptor (NMDAR) family has many critical roles in CNS function and in various neuropathological and psychiatric conditions. Until recently, the types of compounds available to regulate NMDAR function have been quite limited in terms of mechanism of action, subtype selectivity, and biological effect. However, several new classes of NMDAR agents have now been identified that are positive or negative allosteric modulators (PAMs and NAMs, respectively) with various patterns of NMDAR subtype selectivity. These new agents act at several newly recognized binding sites on the NMDAR complex and offer significantly greater pharmacological control over NMDAR activity than previously available agents. The purpose of this review is to summarize the structure-activity relationships for these new NMDAR modulator drug classes and to describe the current understanding of their mechanisms of action.
Figures


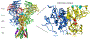

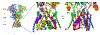






References
-
- Watkins JC; Evans RH Excitatory amino acid transmitters. Annu Rev Pharmacol Toxicol 1981, 21, 165–204. - PubMed
-
- Monaghan DT; Bridges RJ; Cotman CW The excitatory amino acid receptors: their classes, pharmacology, and distinct properties in the function of the central nervous system. Annu Rev Pharmacol Toxicol 1989, 29, 365–402. - PubMed
-
- Dingledine R; Borges K; Bowie D; Traynelis SF The glutamate receptor ion channels. Pharmacol Rev 1999, 51, 7–61. - PubMed
-
- Paoletti P; Bellone C; Zhou Q NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci 2013, 14, 383–400. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

