A comprehensive and quantitative comparison of text-mining in 15 million full-text articles versus their corresponding abstracts
- PMID: 29447159
- PMCID: PMC5831415
- DOI: 10.1371/journal.pcbi.1005962
A comprehensive and quantitative comparison of text-mining in 15 million full-text articles versus their corresponding abstracts
Abstract
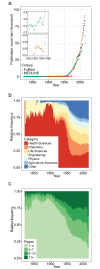
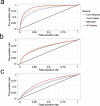
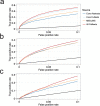
Across academia and industry, text mining has become a popular strategy for keeping up with the rapid growth of the scientific literature. Text mining of the scientific literature has mostly been carried out on collections of abstracts, due to their availability. Here we present an analysis of 15 million English scientific full-text articles published during the period 1823-2016. We describe the development in article length and publication sub-topics during these nearly 250 years. We showcase the potential of text mining by extracting published protein-protein, disease-gene, and protein subcellular associations using a named entity recognition system, and quantitatively report on their accuracy using gold standard benchmark data sets. We subsequently compare the findings to corresponding results obtained on 16.5 million abstracts included in MEDLINE and show that text mining of full-text articles consistently outperforms using abstracts only.
Conflict of interest statement
SB and LJJ are on the scientific advisory board and have been among the founders of Intomics A/S with equity in the company.
Figures
References
-
- Azevedo A. Integration of Data Mining in Business Intelligence Systems 1st Editio Azevedo A, Santos MF, editors. Integration of Data Mining in Business Intelligence Systems. IGI Publishing Hershey, PA, USA; 2014. 314 p.
-
- Krallinger M, Valencia A. Text-mining and information-retrieval services for molecular biology. Vol. 6, Genome biology. 20056(7):224 doi: 10.1186/gb-2005-6-7-224 - DOI - PMC - PubMed
-
- Fleuren WWM, Alkema W. Application of text mining in the biomedical domain. Vol. 74, Methods. 201574:97–106. doi: 10.1016/j.ymeth.2015.01.015 - DOI - PubMed
-
- Luo Y, Riedlinger G, Szolovits P. Text Mining in Cancer Gene and Pathway Prioritization. Vol. 13, Cancer Informatics. 201413(Suppl 1):69–79. doi: 10.4137/CIN.S13874 - DOI - PMC - PubMed
-
- Ananiadou S, Thompson P, Nawaz R, McNaught J, Kell DB. Event-based text mining for biology and functional genomics. Vol. 14, Briefings in functional genomics. 201514(3):213–30. doi: 10.1093/bfgp/elu015 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

