Joint ancestry and association test indicate two distinct pathogenic pathways involved in classical dengue fever and dengue shock syndrome
- PMID: 29447178
- PMCID: PMC5813895
- DOI: 10.1371/journal.pntd.0006202
Joint ancestry and association test indicate two distinct pathogenic pathways involved in classical dengue fever and dengue shock syndrome
Abstract
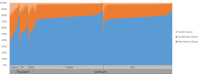
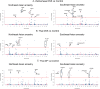
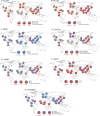
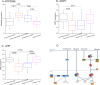
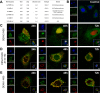
Ethnic diversity has been long considered as one of the factors explaining why the severe forms of dengue are more prevalent in Southeast Asia than anywhere else. Here we take advantage of the admixed profile of Southeast Asians to perform coupled association-admixture analyses in Thai cohorts. For dengue shock syndrome (DSS), the significant haplotypes are located in genes coding for phospholipase C members (PLCB4 added to previously reported PLCE1), related to inflammation of blood vessels. For dengue fever (DF), we found evidence of significant association with CHST10, AHRR, PPP2R5E and GRIP1 genes, which participate in the xenobiotic metabolism signaling pathway. We conducted functional analyses for PPP2R5E, revealing by immunofluorescence imaging that the coded protein co-localizes with both DENV1 and DENV2 NS5 proteins. Interestingly, only DENV2-NS5 migrated to the nucleus, and a deletion of the predicted top-linking motif in NS5 abolished the nuclear transfer. These observations support the existence of differences between serotypes in their cellular dynamics, which may contribute to differential infection outcome risk. The contribution of the identified genes to the genetic risk render Southeast and Northeast Asian populations more susceptible to both phenotypes, while African populations are best protected against DSS and intermediately protected against DF, and Europeans the best protected against DF but the most susceptible against DSS.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–7. Epub 2013/04/09. doi: 10.1038/nature12060 ; PubMed Central PMCID: PMCPmc3651993. - DOI - PMC - PubMed
-
- WHO. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control. 2nd ed. Geneva: World Health Organization; 1997. 84 p.
-
- Khor CC, Chau TN, Pang J, Davila S, Long HT, Ong RT, et al. Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat Genet. 2011;43(11):1139–41. Epub 2011/10/18. doi: 10.1038/ng.960 ; PubMed Central PMCID: PMCPmc3223402. - DOI - PMC - PubMed
-
- Long HT, Hibberd ML, Hien TT, Dung NM, Van Ngoc T, Farrar J, et al. Patterns of gene transcript abundance in the blood of children with severe or uncomplicated dengue highlight differences in disease evolution and host response to dengue virus infection. J Infect Dis. 2009;199(4):537–46. Epub 2009/01/14. doi: 10.1086/596507 ; PubMed Central PMCID: PMCPmc4333209. - DOI - PMC - PubMed
-
- Hoang LT, Lynn DJ, Henn M, Birren BW, Lennon NJ, Le PT, et al. The early whole-blood transcriptional signature of dengue virus and features associated with progression to dengue shock syndrome in Vietnamese children and young adults. Journal of virology. 2010;84(24):12982–94. Epub 2010/10/15. doi: 10.1128/JVI.01224-10 ; PubMed Central PMCID: PMCPmc3004338. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

