Comparing deep learning and concept extraction based methods for patient phenotyping from clinical narratives
- PMID: 29447188
- PMCID: PMC5813927
- DOI: 10.1371/journal.pone.0192360
Comparing deep learning and concept extraction based methods for patient phenotyping from clinical narratives
Abstract
In secondary analysis of electronic health records, a crucial task consists in correctly identifying the patient cohort under investigation. In many cases, the most valuable and relevant information for an accurate classification of medical conditions exist only in clinical narratives. Therefore, it is necessary to use natural language processing (NLP) techniques to extract and evaluate these narratives. The most commonly used approach to this problem relies on extracting a number of clinician-defined medical concepts from text and using machine learning techniques to identify whether a particular patient has a certain condition. However, recent advances in deep learning and NLP enable models to learn a rich representation of (medical) language. Convolutional neural networks (CNN) for text classification can augment the existing techniques by leveraging the representation of language to learn which phrases in a text are relevant for a given medical condition. In this work, we compare concept extraction based methods with CNNs and other commonly used models in NLP in ten phenotyping tasks using 1,610 discharge summaries from the MIMIC-III database. We show that CNNs outperform concept extraction based methods in almost all of the tasks, with an improvement in F1-score of up to 26 and up to 7 percentage points in area under the ROC curve (AUC). We additionally assess the interpretability of both approaches by presenting and evaluating methods that calculate and extract the most salient phrases for a prediction. The results indicate that CNNs are a valid alternative to existing approaches in patient phenotyping and cohort identification, and should be further investigated. Moreover, the deep learning approach presented in this paper can be used to assist clinicians during chart review or support the extraction of billing codes from text by identifying and highlighting relevant phrases for various medical conditions.
Conflict of interest statement
Figures

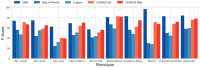
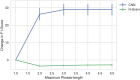
References
-
- Data MC. Secondary Analysis of Electronic Health Records. Springer; 2016. - PubMed
-
- Charles D, Gabriel M, Furukawa MF. Adoption of electronic health record systems among US non-federal acute care hospitals: 2008-2012. ONC data brief. 2013;9:1–9.
-
- Saeed M, Lieu C, Raber G, Mark RG. MIMIC II: a massive temporal ICU patient database to support research in intelligent patient monitoring. In: Computers in Cardiology, 2002. IEEE; 2002. p. 641–644. - PubMed
-
- Johnson AE, Pollard TJ, Shen L, Lehman LwH, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Scientific data. 2016;3 doi: 10.1038/sdata.2016.35 - DOI - PMC - PubMed
-
- Uzuner Ö, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records. Journal of the American Medical Informatics Association. 2008;15(1):14–24. doi: 10.1197/jamia.M2408 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

