Defeating Major Contaminants in Fe3+- Immobilized Metal Ion Affinity Chromatography (IMAC) Phosphopeptide Enrichment
- PMID: 29449344
- PMCID: PMC5930410
- DOI: 10.1074/mcp.TIR117.000518
Defeating Major Contaminants in Fe3+- Immobilized Metal Ion Affinity Chromatography (IMAC) Phosphopeptide Enrichment
Abstract
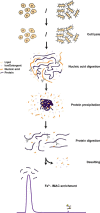
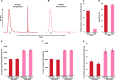
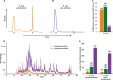
Here we demonstrate that biomolecular contaminants, such as nucleic acid molecules, can seriously interfere with immobilized metal ion affinity chromatography (IMAC)-based phosphopeptide enrichments. We address and largely solve this issue, developing a robust protocol implementing methanol/chloroform protein precipitation and enzymatic digestion using benzonase, which degrades all forms of DNA and RNA, before IMAC-column loading. This simple procedure resulted in a drastic increase of enrichment sensitivity, enabling the identification of around 17,000 unique phosphopeptides and 12,500 unambiguously localized phosphosites in human cell-lines from a single LC-MS/MS run, constituting a 50% increase when compared with the standard protocol. The improved protocol was also applied to bacterial samples, increasing the number of identified bacterial phosphopeptides even more strikingly, by a factor 10, when compared with the standard protocol. For E. coli we detected around 1300 unambiguously localized phosphosites per LC-MS/MS run. The preparation of these ultra-pure phosphopeptide samples only requires marginal extra costs and sample preparation time and should thus be adoptable by every laboratory active in the field of phosphoproteomics.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
References
-
- Hunter T. (2000) Signaling—2000 and Beyond. Cell 100, 113–127 - PubMed
-
- Pawson T. (2004) Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116, 191–203 - PubMed
-
- Ubersax J. A., and Ferrell J. E. Jr (2007) Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530–541 - PubMed
-
- Cohen P. (2001) The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur. J. Biochem. 268, 5001–5010 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

