'Big bang' of B-cell development revealed
- PMID: 29449365
- PMCID: PMC5830931
- DOI: 10.1101/gad.311357.118
'Big bang' of B-cell development revealed
Abstract
Earlier studies have identified transcription factors that specify B-cell fate, but the underlying mechanisms remain to be revealed. Two new studies by Miyai and colleagues (pp. 112-126) and Li and colleagues (pp. 96-111) in this issue of Genes & Development provide new and unprecedented insights into the genetic and epigenetic mechanisms that establish B-cell identity.
Keywords: B-cell differentiation; B-cell programming; EBF1; IRF4; Pax5; chromatin; epigenetics; transcription factor.
© 2018 Murre; Published by Cold Spring Harbor Laboratory Press.
Figures
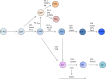
Comment on
-
Three-step transcriptional priming that drives the commitment of multipotent progenitors toward B cells.Genes Dev. 2018 Jan 15;32(2):112-126. doi: 10.1101/gad.309575.117. Epub 2018 Feb 9. Genes Dev. 2018. PMID: 29440259 Free PMC article.
-
Dynamic EBF1 occupancy directs sequential epigenetic and transcriptional events in B-cell programming.Genes Dev. 2018 Jan 15;32(2):96-111. doi: 10.1101/gad.309583.117. Epub 2018 Feb 9. Genes Dev. 2018. PMID: 29440261 Free PMC article.
References
-
- Bain G, Murre C. 1998. The role of E-proteins in B and T-lymphocyte development. Semin Immunol 10: 143–153. - PubMed
-
- Maier H, Ostraat R, Gao H, Fields S, Shinton SA, Medina KL, Ikawa T, Murre C, Singh H, Hardy RR, et al. 2004. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol 5: 1069–1077. - PubMed
-
- Medvedovic J, Ebert A, Tagoh H, Busslinger M. 2011. Pax5: a master regulator of B cell development and leukemogenesis. Adv Immunol 111: 179–206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
