Expression profiling of chromatin-modifying enzymes and global DNA methylation in CD4+ T cells from patients with chronic HIV infection at different HIV control and progression states
- PMID: 29449904
- PMCID: PMC5812196
- DOI: 10.1186/s13148-018-0448-5
Expression profiling of chromatin-modifying enzymes and global DNA methylation in CD4+ T cells from patients with chronic HIV infection at different HIV control and progression states
Abstract
Background: Integration of human immunodeficiency virus type 1 (HIV-1) into the host genome causes global disruption of the chromatin environment. The abundance level of various chromatin-modifying enzymes produces these alterations and affects both the provirus and cellular gene expression. Here, we investigated potential changes in enzyme expression and global DNA methylation in chronically infected individuals with HIV-1 and compared these changes with non-HIV infected individuals. We also evaluated the effect of viral replication and degree of disease progression over these changes.
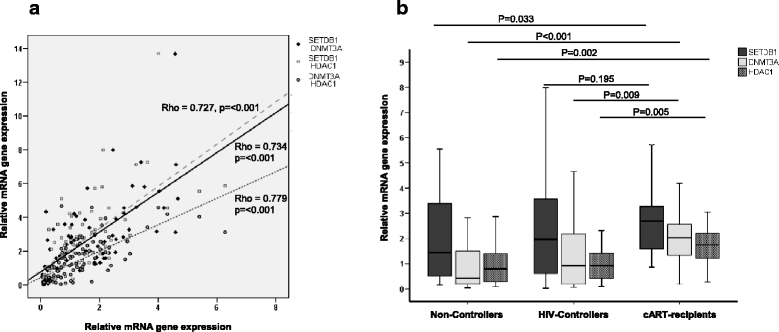
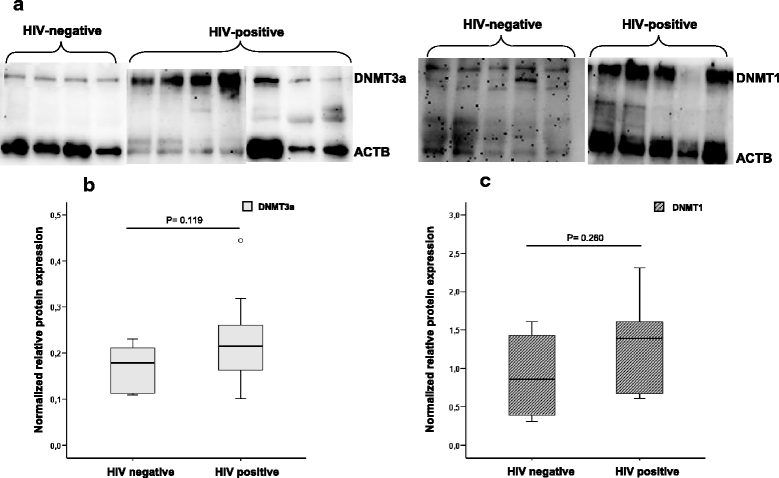
Results: Individuals with HIV-1 had a significant surge in the expression of DNA and histone methyltransferases (DNMT3A and DNMT3B, SETDB1, SUV39H1) compared with non-infected individuals, with the exception of PRMT6, which was downregulated. Some histone deacetylases (HDAC2 and HDAC3) were also upregulated in patients with HIV. Among individuals with HIV-1 with various degrees of progression and HIV control, the group of treated patients with undetectable viremia showed greater differences with the other two groups (untreated HIV-1 controllers and non-controllers). These latter two groups exhibited a similar behavior between them. Of interest, the overexpression of genes that associate with viral protein Tat (such as SETDB1 along with DNMT3A and HDAC1, and SIRT-1) was more prevalent in treated patients. We also observed elevated levels of global DNA methylation in individuals with HIV-1 in an inverse correlation with the CD4/CD8 ratio.
Conclusions: The current study shows an increase in chromatin-modifying enzymes and remodelers and in global DNA methylation in patients with chronic HIV-1 infection, modulated by various levels of viral control and progression.
Keywords: Chromatin-modifying enzymes; DNA methylation; Epigenetics; HDAC; HIV; Methyltransferases; Progression.
Conflict of interest statement
All participating individuals provided their informed written consent, and the protocols were approved by the Hospital Universitario La Paz and Spanish AIDS Research Network ethics committees.Not applicableThe authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Epigenetic Metabolite Acetate Inhibits Class I/II Histone Deacetylases, Promotes Histone Acetylation, and Increases HIV-1 Integration in CD4+ T Cells.J Virol. 2017 Jul 27;91(16):e01943-16. doi: 10.1128/JVI.01943-16. Print 2017 Aug 15. J Virol. 2017. PMID: 28539453 Free PMC article.
-
Productive human immunodeficiency virus type 1 infection in peripheral blood predominantly takes place in CD4/CD8 double-negative T lymphocytes.J Virol. 2007 Sep;81(18):9693-706. doi: 10.1128/JVI.00492-07. Epub 2007 Jul 3. J Virol. 2007. PMID: 17609262 Free PMC article. Clinical Trial.
-
Blimp-1 overexpression is associated with low HIV-1 reservoir and transcription levels in central memory CD4+ T cells from elite controllers.AIDS. 2014 Jul 17;28(11):1567-77. doi: 10.1097/QAD.0000000000000295. AIDS. 2014. PMID: 24804861
-
HIV-Induced Epigenetic Alterations in Host Cells.Adv Exp Med Biol. 2016;879:27-38. doi: 10.1007/978-3-319-24738-0_2. Adv Exp Med Biol. 2016. PMID: 26659262 Review.
-
Chromatin modifications (acetylation/ deacetylation/ methylation) as new targets for HIV therapy.Curr Pharm Des. 2006;12(16):1975-93. doi: 10.2174/138161206777442092. Curr Pharm Des. 2006. PMID: 16787242 Review.
Cited by
-
Relationship between DNA Methylation Profiles and Active Tuberculosis Development from Latent Infection: a Pilot Study in Nested Case-Control Design.Microbiol Spectr. 2022 Jun 29;10(3):e0058622. doi: 10.1128/spectrum.00586-22. Epub 2022 Apr 21. Microbiol Spectr. 2022. PMID: 35446152 Free PMC article.
-
Deciphering a TB-related DNA methylation biomarker and constructing a TB diagnostic classifier.Mol Ther Nucleic Acids. 2021 Nov 19;27:37-49. doi: 10.1016/j.omtn.2021.11.014. eCollection 2022 Mar 8. Mol Ther Nucleic Acids. 2021. PMID: 34938605 Free PMC article.
-
Connection Between HIV and Mitochondria in Cardiovascular Disease and Implications for Treatments.Circ Res. 2024 May 24;134(11):1581-1606. doi: 10.1161/CIRCRESAHA.124.324296. Epub 2024 May 23. Circ Res. 2024. PMID: 38781302 Free PMC article. Review.
-
Impact of chromatin on HIV-1 latency: a multi-dimensional perspective.Epigenetics Chromatin. 2025 Mar 8;18(1):9. doi: 10.1186/s13072-025-00573-x. Epigenetics Chromatin. 2025. PMID: 40055755 Free PMC article. Review.
-
Down-Regulation of the Longevity-Associated Protein SIRT1 in Peripheral Blood Mononuclear Cells of Treated HIV Patients.Cells. 2022 Jan 20;11(3):348. doi: 10.3390/cells11030348. Cells. 2022. PMID: 35159154 Free PMC article.
References
-
- Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, Tyagi M, Karn J. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J Virol. 2008;82:12291–12303. doi: 10.1128/JVI.01383-08. - DOI - PMC - PubMed
-
- Coiras M, López-Huertas MR, Pérez-Olmeda M, Alcamí J. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nature Reviews. 2009;7:798–812. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

