Thermal decomposition of the amino acids glycine, cysteine, aspartic acid, asparagine, glutamic acid, glutamine, arginine and histidine
- PMID: 29449937
- PMCID: PMC5807855
- DOI: 10.1186/s13628-018-0042-4
Thermal decomposition of the amino acids glycine, cysteine, aspartic acid, asparagine, glutamic acid, glutamine, arginine and histidine
Abstract
Background: The pathways of thermal instability of amino acids have been unknown. New mass spectrometric data allow unequivocal quantitative identification of the decomposition products.
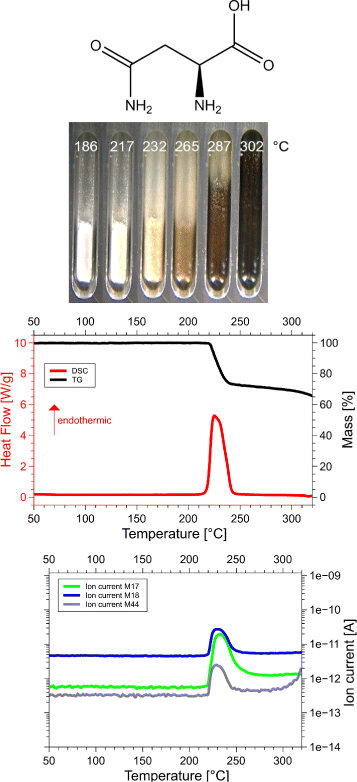
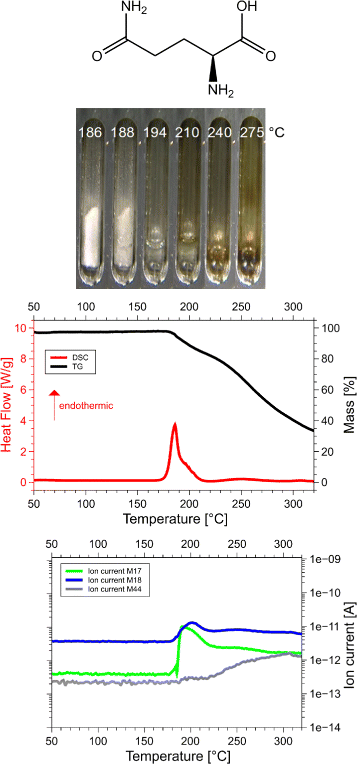
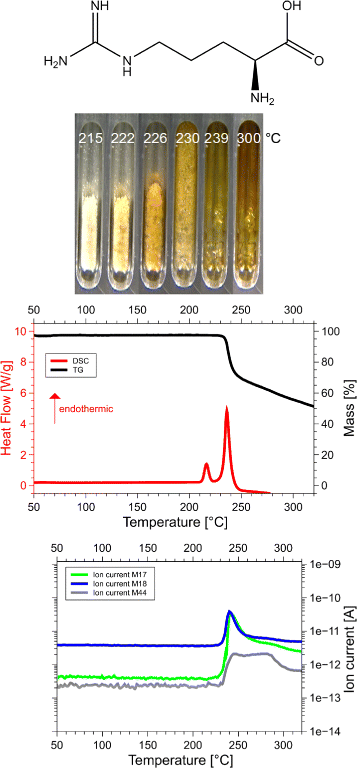
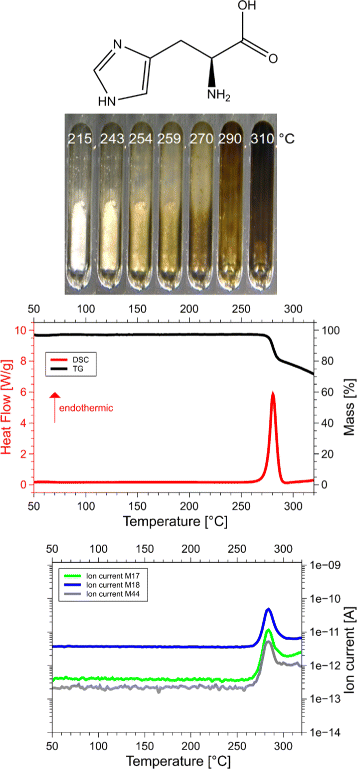
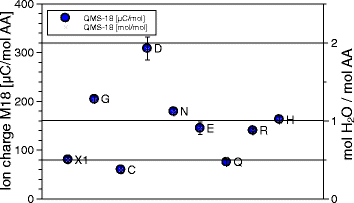
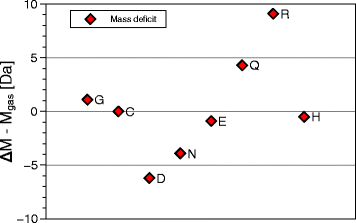
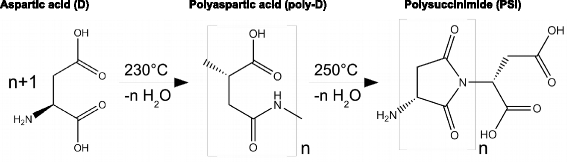
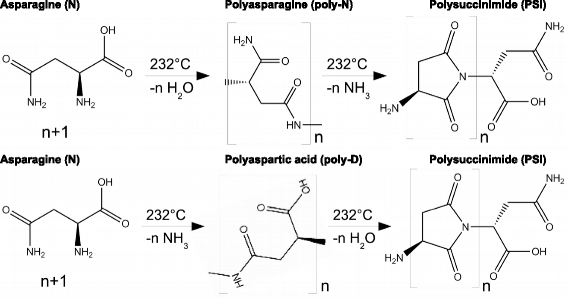
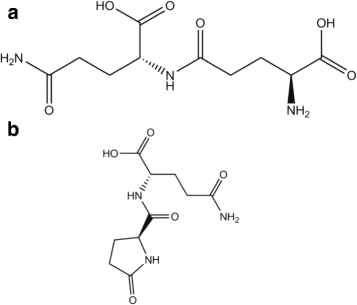
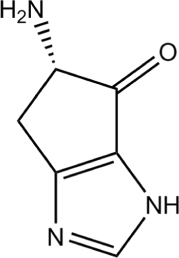
Results: Calorimetry, thermogravimetry and mass spectrometry were used to follow the thermal decomposition of the eight amino acids G, C, D, N, E, Q, R and H between 185 °C and 280 °C. Endothermic heats of decomposition between 72 and 151 kJ/mol are needed to form 12 to 70% volatile products. This process is neither melting nor sublimation. With exception of cysteine they emit mainly H2O, some NH3 and no CO2. Cysteine produces CO2 and little else. The reactions are described by polynomials, AA→a NH3+b H2O+c CO2+d H2S+e residue, with integer or half integer coefficients. The solid monomolecular residues are rich in peptide bonds.
Conclusions: Eight of the 20 standard amino acids decompose at well-defined, characteristic temperatures, in contrast to commonly accepted knowledge. Products of decomposition are simple. The novel quantitative results emphasize the impact of water and cyclic condensates with peptide bonds and put constraints on hypotheses of the origin, state and stability of amino acids in the range between 200 °C and 300 °C.
Keywords: Amino acid; Quantitative mass spectrometry; Thermal analysis.
Conflict of interest statement
Not applicable.Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures











References
-
- Nelson DL, Cox MM. Lehninger Principles of Biochemistry. New York: Macmillan; 2017.
-
- Boldyreva E. Crystalline amino acids. In: Boeyens JCA, Ogilvie JF, editors. Models, Mysteries and Magic of Molecules. Dordrecht: Springer; 2013.
-
- Barret G. Chemistry and Biochemistry of the Amino Acids. Heidelberg: Springer; 1985.
-
- Acree W, Chickos JS. Phase transition enthalpy measurements of organic and organometallic compounds. sublimation, vaporization and fusion enthalpies from 1880 to 2010. J Phys Chem Ref Data. 2010;39:043101. doi: 10.1063/1.3309507. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

