The binding structure and affinity of photodamaged duplex DNA with members of the photolyase/cryptochrome family: A computational study
- PMID: 29450111
- PMCID: PMC5812317
- DOI: 10.2142/biophysico.15.0_18
The binding structure and affinity of photodamaged duplex DNA with members of the photolyase/cryptochrome family: A computational study
Abstract
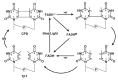
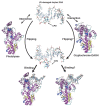
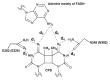
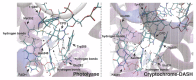
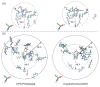
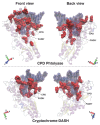
Photolyases (PHRs) and cryptochromes (CRYs) belong to the same family known as blue-light photoreceptors. Although their amino acid sequences and corresponding structures are similar to each other, they exert different functions. PHRs function as an enzyme to repair UV-induced deoxyribonucleic acid (DNA) lesions such as a cyclobutane pyrimidine dimer (CPD) and a (6-4) photoproduct ((6-4)pp), whereas CRYs are a circadian photoreceptor in plants and animals and at the same time they control the photoperiodic induction of flowering in plants. When a new type cryptochrome was identified, it was assumed that another type of CRYs, cryptochrome-DASH (CRY-DASH), which is categorized as a subfamily of photolyase/cryptochrome family, would possess the DNA photolyase activity. However, CRY-DASH had a weak DNA photolyase activity, but the reason for this is still unclear. To clarify the reason, we performed molecular dynamics (MD) simulations for a complex of CPD-PHR or CRY-DASH with damaged double-stranded DNA (dsDNA) and estimated the binding free energy, ΔGbind, between the protein and the damaged dsDNA by using a molecular mechanics/Poisson-Boltzmann surface area (MM/PBSA) method. ΔGbind for both proteins were -35 and 57 kcal mol-1, respectively, indicating that the structural stability of CRY-DASH was lower than that of CPD-PHR upon the damaged dsDNA binding. In particular, the number of amino acid residues relevant to the damaged dsDNA binding on the CRY-DASH surface was smaller than that on CPD-PHR. Therefore, the present result suggests that CRY-DASH has a weak DNA photolyase activity because it has a lower binding affinity than CPD-PHR.
Keywords: binding free energy; duplex DNA binding; molecular mechanics/Poisson-Boltzmann surface area; photolyase/cryptochrome family.
Conflict of interest statement
Conflicts of Interest All the authors declare that they have no conflict of interest.
Figures


Similar articles
-
The DASH-type Cryptochrome from the Fungus Mucor circinelloides Is a Canonical CPD-Photolyase.Curr Biol. 2020 Nov 16;30(22):4483-4490.e4. doi: 10.1016/j.cub.2020.08.051. Epub 2020 Sep 17. Curr Biol. 2020. PMID: 32946746
-
Fungal cryptochrome with DNA repair activity reveals an early stage in cryptochrome evolution.Proc Natl Acad Sci U S A. 2015 Dec 8;112(49):15130-5. doi: 10.1073/pnas.1514637112. Epub 2015 Nov 17. Proc Natl Acad Sci U S A. 2015. PMID: 26578805 Free PMC article.
-
Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity.Proc Natl Acad Sci U S A. 2006 Nov 21;103(47):17701-6. doi: 10.1073/pnas.0608554103. Epub 2006 Nov 13. Proc Natl Acad Sci U S A. 2006. PMID: 17101984 Free PMC article.
-
A Review of Spectroscopic and Biophysical-Chemical Studies of the Complex of Cyclobutane Pyrimidine Dimer Photolyase and Cryptochrome DASH with Substrate DNA.Photochem Photobiol. 2017 Jan;93(1):26-36. doi: 10.1111/php.12678. Epub 2017 Jan 20. Photochem Photobiol. 2017. PMID: 27891613 Review.
-
The Gain and Loss of Cryptochrome/Photolyase Family Members during Evolution.Genes (Basel). 2022 Sep 8;13(9):1613. doi: 10.3390/genes13091613. Genes (Basel). 2022. PMID: 36140781 Free PMC article.
Cited by
-
Theoretical insights into the DNA repair function of Arabidopsis thaliana cryptochrome-DASH.Biophys Physicobiol. 2020 Sep 4;17:113-124. doi: 10.2142/biophysico.BSJ-2020010. eCollection 2020. Biophys Physicobiol. 2020. PMID: 33194514 Free PMC article.
-
Coulomb and CH-π interactions in (6-4) photolyase-DNA complex dominate DNA binding and repair abilities.Nucleic Acids Res. 2018 Jul 27;46(13):6761-6772. doi: 10.1093/nar/gky364. Nucleic Acids Res. 2018. PMID: 29762762 Free PMC article.
References
-
- Taylor J-S. DNA, sunlight, and skin cancer. J Chem Educ. 1990;67:835–841.
-
- Teramura AH. Effects of ultraviolet-B radiation on the growth and yield of crop plants. Physiol Plant. 1983;58:415–427.
-
- de Gruijl FR, Roza L. Photoreactivation in humans. J Photochem Photobiol B, Biol. 1991;10:367–371. - PubMed
-
- Bornman JF, Teramura AH. Effects of ultraviolet-B radiation on terrestrial plants. In: Young AR, Björn LO, Moan J, Nultsch W, editors. Environmental UV-Photobiology. Springer; Boston, MA: 1993. pp. 427–471.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
