Novel proteins from proteomic analysis of the trunk disease fungus Lasiodiplodia theobromae (Botryosphaeriaceae)
- PMID: 29450146
- PMCID: PMC5802045
- DOI: 10.1016/j.biopen.2017.03.001
Novel proteins from proteomic analysis of the trunk disease fungus Lasiodiplodia theobromae (Botryosphaeriaceae)
Abstract
Many basic science questions remain regarding protein functions in the pathogen: host interaction, especially in the trunk disease fungi family, the Botryosphaeriaceae, which are a global problem for economically important plants, especially fruiting trees. Proteomics is a highly useful technology for studying protein expression and for discovering novel proteins in unsequenced and poorly annotated organisms. Current fungal proteomics approaches involve 2D SDS-PAGE and extensive, complex, protein extraction methodologies. In this work, a modified Folch extraction was applied to protein extraction to perform both de novo peptide sequencing and peptide fragmentation analysis/protein identification of the plant and human fungal pathogen Lasiodiplodia theobromae. Both bioinformatics approaches yielded novel peptide sequences from proteins produced by L. theobromae in the presence of exogenous triglycerides and glucose. These proteins and the functions they may possess could be targeted for further functional characterization and validation efforts, due to their potential uses in biotechnology and as new paradigms for understanding fungal biochemistry, such as the finding of allergenic enolases, as well as various novel proteases, including zinc metalloproteinases homologous to those found in snake venom. This work contributes to genomic annotation efforts, which, hand in hand with genomic sequencing, will help improve fungal bioinformatics databases for future studies of Botryosphaeriaceae. All data, including raw data, are available via the ProteomeXchange data repository with identifier PXD005283. This is the first study of its kind in Botryosphaeriaceae.
Keywords: Bioinformatics; Gene ontology; Peptide fragmentation analysis; Trunk-disease fungi; de novo peptide sequencing.
Figures
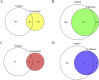
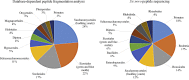
References
-
- Blackwell M. The fungi: 1, 2, 3... 5.1 million species? Am. J. Bot. 2011;98(3):426–438. - PubMed
-
- Brambl R. Fungal physiology and the origins of molecular biology. Microbiology. 2009;155(12):3799–3809. - PubMed
-
- Carnielli C.M., Winck F.V., Paes Leme A.F. Functional annotation and biological interpretation of proteomics data. Biochim. Biophys. Acta – Proteins Proteomics. 2015;1854(1):46–54. - PubMed
-
- Ambrogelly A., Palioura S., Söll D. Natural expansion of the genetic code. Nat. Chem. Biol. 2007;3(1):29–35. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

