Can dimedone be used to study selenoproteins? An investigation into the reactivity of dimedone toward oxidized forms of selenocysteine
- PMID: 29451338
- PMCID: PMC6295895
- DOI: 10.1002/pro.3390
Can dimedone be used to study selenoproteins? An investigation into the reactivity of dimedone toward oxidized forms of selenocysteine
Abstract
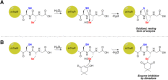
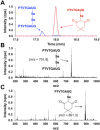
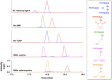
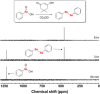
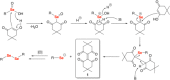
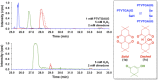
Dimedone is a widely used reagent to assess the redox state of cysteine-containing proteins as it will alkylate sulfenic acid residues, but not sulfinic acid residues. While it has been reported that dimedone can label selenenic acid residues in selenoproteins, we investigated the stability, and reversibility of this label in a model peptide system. We also wondered whether dimedone could be used to detect seleninic acid residues. We used benzenesulfinic acid, benzeneseleninic acid, and model selenocysteine-containing peptides to investigate possible reactions with dimedone. These peptides were incubated with H2 O2 in the presence of dimedone and then the reactions were followed by liquid chromatography/electrospray ionization mass spectrometry (LC/ESI-MS). The native peptide, H-PTVTGCUG-OH (corresponding to the native amino acid sequence of the C-terminus of mammalian thioredoxin reductase), could not be alkylated by dimedone, but could be carboxymethylated with iodoacetic acid. However the "mutant peptide," H-PTVTGAUG-OH, could be labeled with dimedone at low concentrations of H2 O2 , but the reaction was reversible by addition of thiol. Due to the reversible nature of this alkylation, we conclude that dimedone is not a good reagent for detecting selenenic acids in selenoproteins. At high concentrations of H2 O2 , selenium was eliminated from the peptide and a dimeric form of dimedone could be detected using LCMS and 1 H NMR. The dimeric dimedone product forms as a result of a seleno-Pummerer reaction with Sec-seleninic acid. Overall our results show that the reaction of dimedone with oxidized cysteine residues is quite different from the same reaction with oxidized selenocysteine residues.
Keywords: Pummerer reaction; dimedone; selenenic acid; seleninic acid; selenocysteine; sulfenic acid; sulfinic acid.
© 2018 The Protein Society.
Figures

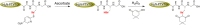



References
-
- Reich HJ, Hondal RJ (2016) Why nature chose selenium. ACS Chem Biol 11:821–841. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

