A stochastic and dynamical view of pluripotency in mouse embryonic stem cells
- PMID: 29451874
- PMCID: PMC5833290
- DOI: 10.1371/journal.pcbi.1006000
A stochastic and dynamical view of pluripotency in mouse embryonic stem cells
Abstract
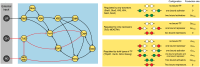
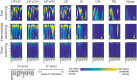
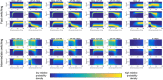
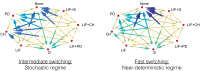
Pluripotent embryonic stem cells are of paramount importance for biomedical sciences because of their innate ability for self-renewal and differentiation into all major cell lines. The fateful decision to exit or remain in the pluripotent state is regulated by complex genetic regulatory networks. The rapid growth of single-cell sequencing data has greatly stimulated applications of statistical and machine learning methods for inferring topologies of pluripotency regulating genetic networks. The inferred network topologies, however, often only encode Boolean information while remaining silent about the roles of dynamics and molecular stochasticity inherent in gene expression. Herein we develop a framework for systematically extending Boolean-level network topologies into higher resolution models of networks which explicitly account for the promoter architectures and gene state switching dynamics. We show the framework to be useful for disentangling the various contributions that gene switching, external signaling, and network topology make to the global heterogeneity and dynamics of transcription factor populations. We find the pluripotent state of the network to be a steady state which is robust to global variations of gene switching rates which we argue are a good proxy for epigenetic states of individual promoters. The temporal dynamics of exiting the pluripotent state, on the other hand, is significantly influenced by the rates of genetic switching which makes cells more responsive to changes in extracellular signals.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures


References
-
- Evans M. Discovering pluripotency: 30 years of mouse embryonic stem cells. Nat Rev Mol Cell Biol. 2011;12(10):680–686. doi: 10.1038/nrm3190 - DOI - PubMed
-
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132(4):661–680. - PubMed
-
- Martello G, Smith A. The nature of embryonic stem cells. Ann Rev Cell Dev Biol. 2014;30(1):647 doi: 10.1146/annurev-cellbio-100913-013116 - DOI - PubMed
-
- Van Kampen NG. Stochastic processes in physics and chemistry. vol. 1 Elsevier; 1992.
-
- Symmons O, Raj A. What’s Luck Got to Do with It: Single Cells, Multiple Fates, and Biological Nondeterminism. Mol cell. 2016;62(5):788–802. doi: 10.1016/j.molcel.2016.05.023 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

