Comparative genomic analysis of the multispecies probiotic-marketed product VSL#3
- PMID: 29451876
- PMCID: PMC5815585
- DOI: 10.1371/journal.pone.0192452
Comparative genomic analysis of the multispecies probiotic-marketed product VSL#3
Erratum in
-
Correction: Comparative genomic analysis of the multispecies probiotic-marketed product VSL#3.PLoS One. 2018 Aug 30;13(8):e0203548. doi: 10.1371/journal.pone.0203548. eCollection 2018. PLoS One. 2018. PMID: 30161239 Free PMC article.
Abstract
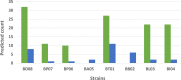
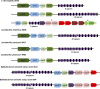
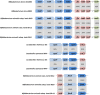
Several probiotic-marketed formulations available for the consumers contain live lactic acid bacteria and/or bifidobacteria. The multispecies product commercialized as VSL#3 has been used for treating various gastro-intestinal disorders. However, like many other products, the bacterial strains present in VSL#3 have only been characterized to a limited extent and their efficacy as well as their predicted mode of action remain unclear, preventing further applications or comparative studies. In this work, the genomes of all eight bacterial strains present in VSL#3 were sequenced and characterized, to advance insights into the possible mode of action of this product and also to serve as a basis for future work and trials. Phylogenetic and genomic data analysis allowed us to identify the 7 species present in the VSL#3 product as specified by the manufacturer. The 8 strains present belong to the species Streptococcus thermophilus, Lactobacillus acidophilus, Lactobacillus paracasei, Lactobacillus plantarum, Lactobacillus helveticus, Bifidobacterium breve and B. animalis subsp. lactis (two distinct strains). Comparative genomics revealed that the draft genomes of the S. thermophilus and L. helveticus strains were predicted to encode most of the defence systems such as restriction modification and CRISPR-Cas systems. Genes associated with a variety of potential probiotic functions were also identified. Thus, in the three Bifidobacterium spp., gene clusters were predicted to encode tight adherence pili, known to promote bacteria-host interaction and intestinal barrier integrity, and to impact host cell development. Various repertoires of putative signalling proteins were predicted to be encoded by the genomes of the Lactobacillus spp., i.e. surface layer proteins, LPXTG-containing proteins, or sortase-dependent pili that may interact with the intestinal mucosa and dendritic cells. Taken altogether, the individual genomic characterization of the strains present in the VSL#3 product confirmed the product specifications, determined its coding capacity as well as identified potential probiotic functions.
Conflict of interest statement
Figures

References
-
- Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, et al. (2014) Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11: 506–514. doi: 10.1038/nrgastro.2014.66 - DOI - PubMed
-
- Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, et al. (2009) Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc Natl Acad Sci U S A 106: 17193–17198. doi: 10.1073/pnas.0908876106 - DOI - PMC - PubMed
-
- Douillard FP, Ribbera A, Kant R, Pietilä TE, Järvinen HM, et al. (2013) Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet 9: e1003683 doi: 10.1371/journal.pgen.1003683 - DOI - PMC - PubMed
-
- Nissilä E, Douillard FP, Ritari J, Paulin L, Järvinen HM, et al. (2017) Genotypic and phenotypic diversity of Lactobacillus rhamnosus clinical isolates, their comparison with strain GG and their recognition by complement system. PLoS One 12: e0176739 doi: 10.1371/journal.pone.0176739 - DOI - PMC - PubMed
-
- Tytgat HL, Douillard FP, Reunanen J, Rasinkangas P, Hendrickx AP, et al. (2016) Lactobacillus rhamnosus GG outcompetes Enterococcus faecium via mucus-binding pili: Evidence for a novel and heterospecific probiotic mechanism. Appl Environ Microbiol 82: 5756–5762. doi: 10.1128/AEM.01243-16 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

