Trade-off between synergy and efficacy in combinations of HIV-1 latency-reversing agents
- PMID: 29451894
- PMCID: PMC5833289
- DOI: 10.1371/journal.pcbi.1006004
Trade-off between synergy and efficacy in combinations of HIV-1 latency-reversing agents
Abstract
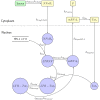
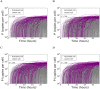
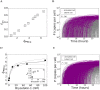
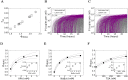
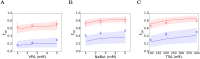
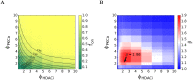
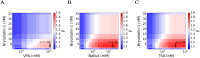
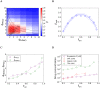
Eradicating HIV-1 infection is difficult because of the reservoir of latently infected cells that gets established soon after infection, remains hidden from antiretroviral drugs and host immune responses, and retains the capacity to reignite infection following the cessation of treatment. Drugs called latency-reversing agents (LRAs) are being developed to reactivate latently infected cells and render them susceptible to viral cytopathicity or immune killing. Whereas individual LRAs have failed to induce adequate reactivation, pairs of LRAs have been identified recently that act synergistically and hugely increase reactivation levels compared to individual LRAs. The maximum synergy achievable with LRA pairs is of clinical importance, as it would allow latency-reversal with minimal drug exposure. Here, we employed stochastic simulations of HIV-1 transcription and translation in latently infected cells to estimate this maximum synergy. We incorporated the predominant mechanisms of action of the two most promising classes of LRAs, namely, protein kinase C agonists and histone deacetylase inhibitors, and quantified the activity of individual LRAs in the two classes by mapping our simulations to corresponding in vitro experiments. Without any adjustable parameters, our simulations then quantitatively captured experimental observations of latency-reversal when the LRAs were used in pairs. Performing simulations representing a wide range of drug concentrations, we estimated the maximum synergy achievable with these LRA pairs. Importantly, we found with all the LRA pairs we considered that concentrations yielding the maximum synergy did not yield the maximum latency-reversal. Increasing concentrations to increase latency-reversal compromised synergy, unravelling a trade-off between synergy and efficacy in LRA combinations. The maximum synergy realizable with LRA pairs would thus be restricted by the desired level of latency-reversal, a constrained optimum we elucidated with our simulations. We expect this trade-off to be important in defining optimal LRA combinations that would maximize synergy while ensuring adequate latency-reversal.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Archin NM, Sung JM, Garrido C, Soriano-Sarabia N, Margolis DM. Eradicating HIV-1 infection: seeking to clear a persistent pathogen. Nat Rev Microbiol. 2014;12:750–64. doi: 10.1038/nrmicro3352 - DOI - PMC - PubMed
-
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–7. doi: 10.1038/8394 - DOI - PubMed
-
- Joos B, Fischer M, Kuster H, Pillai SK, Wong JK, Boni J, et al. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci U S A. 2008;105:16725–30. doi: 10.1073/pnas.0804192105 - DOI - PMC - PubMed
-
- Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–51. doi: 10.1016/j.cell.2013.09.020 - DOI - PMC - PubMed
-
- Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature. 2014;512:74–7. doi: 10.1038/nature13594 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

