An upstream sequence modulates phenazine production at the level of transcription and translation in the biological control strain Pseudomonas chlororaphis 30-84
- PMID: 29451920
- PMCID: PMC5815613
- DOI: 10.1371/journal.pone.0193063
An upstream sequence modulates phenazine production at the level of transcription and translation in the biological control strain Pseudomonas chlororaphis 30-84
Abstract
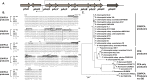
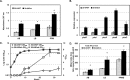
Phenazines are bacterial secondary metabolites and play important roles in the antagonistic activity of the biological control strain P. chlororaphis 30-84 against take-all disease of wheat. The expression of the P. chlororaphis 30-84 phenazine biosynthetic operon (phzXYFABCD) is dependent on the PhzR/PhzI quorum sensing system located immediately upstream of the biosynthetic operon as well as other regulatory systems including Gac/Rsm. Bioinformatic analysis of the sequence between the divergently oriented phzR and phzX promoters identified features within the 5'-untranslated region (5'-UTR) of phzX that are conserved only among 2OHPCA producing Pseudomonas. The conserved sequence features are potentially capable of producing secondary structures that negatively modulate one or both promoters. Transcriptional and translational fusion assays revealed that deletion of 90-bp of sequence at the 5'-UTR of phzX led to up to 4-fold greater expression of the reporters with the deletion compared to the controls, which indicated this sequence negatively modulates phenazine gene expression both transcriptionally and translationally. This 90-bp sequence was deleted from the P. chlororaphis 30-84 chromosome, resulting in 30-84Enh, which produces significantly more phenazine than the wild-type while retaining quorum sensing control. The transcriptional expression of phzR/phzI and amount of AHL signal produced by 30-84Enh also were significantly greater than for the wild-type, suggesting this 90-bp sequence also negatively affects expression of the quorum sensing genes. In addition, deletion of the 90-bp partially relieved RsmE-mediated translational repression, indicating a role for Gac/RsmE interaction. Compared to the wild-type, enhanced phenazine production by 30-84Enh resulted in improvement in fungal inhibition, biofilm formation, extracellular DNA release and suppression of take-all disease of wheat in soil without negative consequences on growth or rhizosphere persistence. This work provides greater insight into the regulation of phenazine biosynthesis with potential applications for improved biological control.
Conflict of interest statement
Figures



References
-
- Pierson LS III, Thomashow LS. (1992) Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30–84. Mol Plant-Microbe Interact.5:330–9. - PubMed
-
- Chin-A-Woeng TF, Bloemberg GV, van der Bij AJ, van der Drift KM, Schripsema J, Kroon B, et al. (1998) Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant-Microbe Interact.11:1069–77.
-
- Powell J, Vargas J Jr, Nair M, Detweiler A, Chandra A. (2000) Management of dollar spot on creeping bentgrass with metabolites of Pseudomonas aureofaciens (TX-1). Plant Dis.84:19–24. - PubMed
-
- Tambong JT, Höfte M. (2001) Phenazines are involved in biocontrol of Pythium myriotylum on cocoyam by Pseudomonas aeruginosa PNA1. Eur J Plant Pathol.107:511–21.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

