Convergent Amino Acid Signatures in Polyphyletic Campylobacter jejuni Subpopulations Suggest Human Niche Tropism
- PMID: 29452359
- PMCID: PMC5841378
- DOI: 10.1093/gbe/evy026
Convergent Amino Acid Signatures in Polyphyletic Campylobacter jejuni Subpopulations Suggest Human Niche Tropism
Abstract
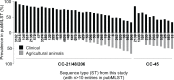
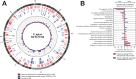
Human infection with the gastrointestinal pathogen Campylobacter jejuni is dependent upon the opportunity for zoonotic transmission and the ability of strains to colonize the human host. Certain lineages of this diverse organism are more common in human infection but the factors underlying this overrepresentation are not fully understood. We analyzed 601 isolate genomes from agricultural animals and human clinical cases, including isolates from the multihost (ecological generalist) ST-21 and ST-45 clonal complexes (CCs). Combined nucleotide and amino acid sequence analysis identified 12 human-only amino acid KPAX clusters among polyphyletic lineages within the common disease causing CC21 group isolates, with no such clusters among CC45 isolates. Isolate sequence types within human-only CC21 group KPAX clusters have been sampled from other hosts, including poultry, so rather than representing unsampled reservoir hosts, the increase in relative frequency in human infection potentially reflects a genetic bottleneck at the point of human infection. Consistent with this, sequence enrichment analysis identified nucleotide variation in genes with putative functions related to human colonization and pathogenesis, in human-only clusters. Furthermore, the tight clustering and polyphyly of human-only lineage clusters within a single CC suggest the repeated evolution of human association through acquisition of genetic elements within this complex. Taken together, combined nucleotide and amino acid analysis of large isolate collections may provide clues about human niche tropism and the nature of the forces that promote the emergence of clinically important C. jejuni lineages.
Figures

References
-
- Asakura H, Yamasaki M, Yamamoto S, Igimi S.. 2007. Deletion of peb4 gene impairs cell adhesion and biofilm formation in Campylobacter jejuni. FEMS Microbiol Lett. 2752:278–285. - PubMed
-
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 195:455–477.http://dx.doi.org/10.1089/cmb.2012.0021 - DOI - PMC - PubMed
-
- Bayliss CD, et al. 2012. Phase variable genes of Campylobacter jejuni exhibit high mutation rates and specific mutational patterns but mutability is not the major determinant of population structure during host colonization. Nucleic Acids Res. 4013:5876–5889.http://dx.doi.org/10.1093/nar/gks246 - DOI - PMC - PubMed
-
- Bentley SD, Parkhill J.. 2004. Comparative genomic structure of prokaryotes. Annu Rev Genet. 38:771–792.http://dx.doi.org/10.1146/annurev.genet.38.072902.094318 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

