Mitochondrial MDM2 Regulates Respiratory Complex I Activity Independently of p53
- PMID: 29452639
- PMCID: PMC8215449
- DOI: 10.1016/j.molcel.2018.01.023
Mitochondrial MDM2 Regulates Respiratory Complex I Activity Independently of p53
Abstract
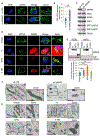
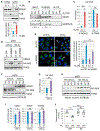
Accumulating evidence indicates that the MDM2 oncoprotein promotes tumorigenesis beyond its canonical negative effects on the p53 tumor suppressor, but these p53-independent functions remain poorly understood. Here, we show that a fraction of endogenous MDM2 is actively imported in mitochondria to control respiration and mitochondrial dynamics independently of p53. Mitochondrial MDM2 represses the transcription of NADH-dehydrogenase 6 (MT-ND6) in vitro and in vivo, impinging on respiratory complex I activity and enhancing mitochondrial ROS production. Recruitment of MDM2 to mitochondria increases during oxidative stress and hypoxia. Accordingly, mice lacking MDM2 in skeletal muscles exhibit higher MT-ND6 levels, enhanced complex I activity, and increased muscular endurance in mild hypoxic conditions. Furthermore, increased mitochondrial MDM2 levels enhance the migratory and invasive properties of cancer cells. Collectively, these data uncover a previously unsuspected function of the MDM2 oncoprotein in mitochondria that play critical roles in skeletal muscle physiology and may contribute to tumor progression.
Keywords: MDM2; MT-ND6; hypoxia; migration; mitochondria; respiratory complex I.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
Figures





References
-
- Ahn BY, Trinh DLN, Zajchowski LD, Lee B, Elwi AN, and Kim SW (2010). Tid1 is a new regulator of p53 mitochondrial translocation and apoptosis in cancer. Oncogene 29, 1155–1166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

