Phasevarions of Bacterial Pathogens: Methylomics Sheds New Light on Old Enemies
- PMID: 29452952
- PMCID: PMC6054543
- DOI: 10.1016/j.tim.2018.01.008
Phasevarions of Bacterial Pathogens: Methylomics Sheds New Light on Old Enemies
Abstract
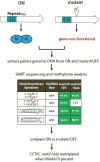
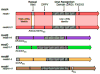
A wide variety of bacterial pathogens express phase-variable DNA methyltransferases that control expression of multiple genes via epigenetic mechanisms. These randomly switching regulons - phasevarions - regulate genes involved in pathogenesis, host adaptation, and antibiotic resistance. Individual phase-variable genes can be identified in silico as they contain easily recognized features such as simple sequence repeats (SSRs) or inverted repeats (IRs) that mediate the random switching of expression. Conversely, phasevarion-controlled genes do not contain any easily identifiable features. The study of DNA methyltransferase specificity using Single-Molecule, Real-Time (SMRT) sequencing and methylome analysis has rapidly advanced the analysis of phasevarions by allowing methylomics to be combined with whole-transcriptome/proteome analysis to comprehensively characterize these systems in a number of important bacterial pathogens.
Keywords: DNA methyltransferase; SMRT sequencing; methylome analysis; phase variation; phasevarion.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures

References
-
- Adhikari S, Curtis PD. DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol Rev. 2016;40:575–591. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

