Physical basis of amyloid fibril polymorphism
- PMID: 29453354
- PMCID: PMC5816019
- DOI: 10.1038/s41467-018-03164-5
Physical basis of amyloid fibril polymorphism
Abstract
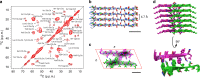
Polymorphism is a key feature of amyloid fibril structures but it remains challenging to explain these variations for a particular sample. Here, we report electron cryomicroscopy-based reconstructions from different fibril morphologies formed by a peptide fragment from an amyloidogenic immunoglobulin light chain. The observed fibril morphologies vary in the number and cross-sectional arrangement of a structurally conserved building block. A comparison with the theoretically possible constellations reveals the experimentally observed spectrum of fibril morphologies to be governed by opposing sets of forces that primarily arise from the β-sheet twist, as well as peptide-peptide interactions within the fibril cross-section. Our results provide a framework for rationalizing and predicting the structure and polymorphism of cross-β fibrils, and suggest that a small number of physical parameters control the observed fibril architectures.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

