Next-generation metabolic screening: targeted and untargeted metabolomics for the diagnosis of inborn errors of metabolism in individual patients
- PMID: 29453510
- PMCID: PMC5959972
- DOI: 10.1007/s10545-017-0131-6
Next-generation metabolic screening: targeted and untargeted metabolomics for the diagnosis of inborn errors of metabolism in individual patients
Abstract
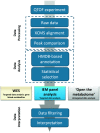
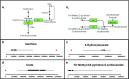
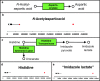
The implementation of whole-exome sequencing in clinical diagnostics has generated a need for functional evaluation of genetic variants. In the field of inborn errors of metabolism (IEM), a diverse spectrum of targeted biochemical assays is employed to analyze a limited amount of metabolites. We now present a single-platform, high-resolution liquid chromatography quadrupole time of flight (LC-QTOF) method that can be applied for holistic metabolic profiling in plasma of individual IEM-suspected patients. This method, which we termed "next-generation metabolic screening" (NGMS), can detect >10,000 features in each sample. In the NGMS workflow, features identified in patient and control samples are aligned using the "various forms of chromatography mass spectrometry (XCMS)" software package. Subsequently, all features are annotated using the Human Metabolome Database, and statistical testing is performed to identify significantly perturbed metabolite concentrations in a patient sample compared with controls. We propose three main modalities to analyze complex, untargeted metabolomics data. First, a targeted evaluation can be done based on identified genetic variants of uncertain significance in metabolic pathways. Second, we developed a panel of IEM-related metabolites to filter untargeted metabolomics data. Based on this IEM-panel approach, we provided the correct diagnosis for 42 of 46 IEMs. As a last modality, metabolomics data can be analyzed in an untargeted setting, which we term "open the metabolome" analysis. This approach identifies potential novel biomarkers in known IEMs and leads to identification of biomarkers for as yet unknown IEMs. We are convinced that NGMS is the way forward in laboratory diagnostics of IEMs.
Keywords: Biomarkers; Canavan disease; High-resolution; Inborn errors of metabolism; Innovative laboratory diagnostics; Mass spectrometry; Metabolomics; QTOF; Xanthinuria.
Conflict of interest statement
Conflict of interest
K. L. M. Coene, L. A. J. Kluijtmans, E. van der Heeft, U. F. H. Engelke, S. de Boer, B. Hoegen, H. J. T. Kwast, M. van de Vorst, M. C. D. G. Huigen, I. M. L.W. Keularts, M. F. Schreuder, C. D. M. van Karnebeek, S. B. Wortmann, M. C. de Vries, M. C. H. Janssen, C. Gilissen, J. Engel and R. A. Wevers declare that they have no conflict of interest.
Informed consent
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained from all patients (or their legal guardians) for being included in the study.
Figures

References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57(1):289–300.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

