AOP-DB: A database resource for the exploration of Adverse Outcome Pathways through integrated association networks
- PMID: 29454060
- PMCID: PMC6104822
- DOI: 10.1016/j.taap.2018.02.006
AOP-DB: A database resource for the exploration of Adverse Outcome Pathways through integrated association networks
Abstract
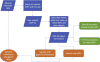
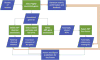
The Adverse Outcome Pathway (AOP) framework describes the progression of a toxicity pathway from molecular perturbation to population-level outcome in a series of measurable, mechanistic responses. The controlled, computer-readable vocabulary that defines an AOP has the ability to, automatically and on a large scale, integrate AOP knowledge with publically available sources of biological high-throughput data and its derived associations. To support the discovery and development of putative (existing) and potential AOPs, we introduce the AOP-DB, an exploratory database resource that aggregates association relationships between genes and their related chemicals, diseases, pathways, species orthology information, ontologies, and gene interactions. These associations are mined from publically available annotation databases and are integrated with the AOP information centralized in the AOP-Wiki, allowing for the automatic characterization of both putative and potential AOPs in the context of multiple areas of biological information, referred to here as "biological entities". The AOP-DB acts as a hypothesis-generation tool for the expansion of putative AOPs, as well as the characterization of potential AOPs, through the creation of association networks across these biological entities. Finally, the AOP-DB provides a useful interface between the AOP framework and existing chemical screening and prioritization efforts by the US Environmental Protection Agency.
Keywords: Adverse Outcome Pathways (AOPs); Association networks; Data integration; Data mining; Database; Risk assessment.
Published by Elsevier Inc.
Figures




References
-
- Ahmed GM, Davies DR. Chronic organophosphate exposure: toward the definition of a neuropsychiatric syndrome. Journal of Nutritional and Environmental Medicine. 2009;7:169–176. doi: 10.1080/13590849762583. - DOI
-
- Ankley GT, Jensen KM, Makynen EA, Kahl MD, Korte JJ, Hornung MW, Gray LE. Effects of the androgenic growth promoter 17-beta-trenbolone on fecundity and reproductive endocrinology of the fathead minnow. Environ Toxicol Chem. 2003;22(6):1350–1360. - PubMed
-
- Arun M, Palimar V. Neurological manifestations in Organophosphorous toxicity. Journal of Indian Academic Forensic Medicine. 2005;30:29–31.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

