Exploring autophagy with Gene Ontology
- PMID: 29455577
- PMCID: PMC5915032
- DOI: 10.1080/15548627.2017.1415189
Exploring autophagy with Gene Ontology
Abstract
Autophagy is a fundamental cellular process that is well conserved among eukaryotes. It is one of the strategies that cells use to catabolize substances in a controlled way. Autophagy is used for recycling cellular components, responding to cellular stresses and ridding cells of foreign material. Perturbations in autophagy have been implicated in a number of pathological conditions such as neurodegeneration, cardiac disease and cancer. The growing knowledge about autophagic mechanisms needs to be collected in a computable and shareable format to allow its use in data representation and interpretation. The Gene Ontology (GO) is a freely available resource that describes how and where gene products function in biological systems. It consists of 3 interrelated structured vocabularies that outline what gene products do at the biochemical level, where they act in a cell and the overall biological objectives to which their actions contribute. It also consists of 'annotations' that associate gene products with the terms. Here we describe how we represent autophagy in GO, how we create and define terms relevant to autophagy researchers and how we interrelate those terms to generate a coherent view of the process, therefore allowing an interoperable description of its biological aspects. We also describe how annotation of gene products with GO terms improves data analysis and interpretation, hence bringing a significant benefit to this field of study.
Keywords: Gene Ontology; Parkinson disease; annotation; autophagy; biocuration; curation; enrichment analysis; network analysis.
Figures

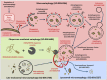

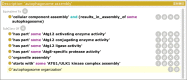

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
