Circadian pathway genetic variation and cancer risk: evidence from genome-wide association studies
- PMID: 29455641
- PMCID: PMC5817863
- DOI: 10.1186/s12916-018-1010-1
Circadian pathway genetic variation and cancer risk: evidence from genome-wide association studies
Abstract
Background: Dysfunction of the circadian clock and single polymorphisms of some circadian genes have been linked to cancer susceptibility, although data are scarce and findings inconsistent. We aimed to investigate the association between circadian pathway genetic variation and risk of developing common cancers based on the findings of genome-wide association studies (GWASs).
Methods: Single nucleotide polymorphisms (SNPs) of 17 circadian genes reported by three GWAS meta-analyses dedicated to breast (Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) Consortium; cases, n = 15,748; controls, n = 18,084), prostate (Elucidating Loci Involved in Prostate Cancer Susceptibility (ELLIPSE) Consortium; cases, n = 14,160; controls, n = 12,724) and lung carcinoma (Transdisciplinary Research In Cancer of the Lung (TRICL) Consortium; cases, n = 12,160; controls, n = 16,838) in patients of European ancestry were utilized to perform pathway analysis by means of the adaptive rank truncated product (ARTP) method. Data were also available for the following subgroups: estrogen receptor negative breast cancer, aggressive prostate cancer, squamous lung carcinoma and lung adenocarcinoma.
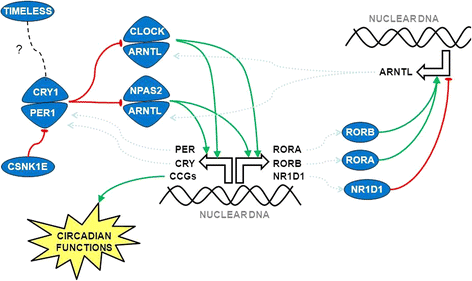
Results: We found a highly significant statistical association between circadian pathway genetic variation and the risk of breast (pathway P value = 1.9 × 10-6; top gene RORA, gene P value = 0.0003), prostate (pathway P value = 4.1 × 10-6; top gene ARNTL, gene P value = 0.0002) and lung cancer (pathway P value = 6.9 × 10-7; top gene RORA, gene P value = 2.0 × 10-6), as well as all their subgroups. Out of 17 genes investigated, 15 were found to be significantly associated with the risk of cancer: four genes were shared by all three malignancies (ARNTL, CLOCK, RORA and RORB), two by breast and lung cancer (CRY1 and CRY2) and three by prostate and lung cancer (NPAS2, NR1D1 and PER3), whereas four genes were specific for lung cancer (ARNTL2, CSNK1E, NR1D2 and PER2) and two for breast cancer (PER1, RORC).
Conclusions: Our findings, based on the largest series ever utilized for ARTP-based gene and pathway analysis, support the hypothesis that circadian pathway genetic variation is involved in cancer predisposition.
Keywords: Cancer predisposition; Cancer risk; Circadian clock; Gene pathway; Genetic variation; Genome-wide association study (GWAS); Germline; Pathway analysis; Single nucleotide polymorphism (SNP).
Conflict of interest statement
Ethics approval and consent to participate
No ethical committee approval was required.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

