The Unusual Genetics and Biochemistry of Bovine Immunoglobulins
- PMID: 29455846
- PMCID: PMC5935254
- DOI: 10.1016/bs.ai.2017.12.004
The Unusual Genetics and Biochemistry of Bovine Immunoglobulins
Abstract
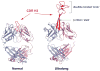
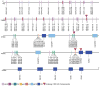
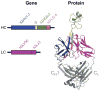
Antibodies are the key circulating molecules that have evolved to fight infection by the adaptive immune system of vertebrates. Typical antibodies of most species contain six complementarity-determining regions (CDRs), where the third CDR of the heavy chain (CDR H3) has the greatest diversity and often makes the most significant contact with antigen. Generally, the process of V(D)J recombination produces a vast repertoire of antibodies; multiple V, D, and J gene segments recombine with additional junctional diversity at the V-D and D-J joints, and additional combinatorial possibilities occur through heavy- and light-chain pairing. Despite these processes, the overall structure of the resulting antibody is largely conserved, and binding to antigen occurs predominantly through the CDR loops of the immunoglobulin V domains. Bovines have deviated from this general paradigm by having few VH regions and thus little germline combinatorial diversity, but their antibodies contain long CDR H3 regions, with substantial diversity generated through somatic hypermutation. A subset of the repertoire comprises antibodies with ultralong CDR H3s, which can reach over 70 amino acids in length. Structurally, these unusual antibodies form a β-ribbon "stalk" and disulfide-bonded "knob" that protrude far from the antibody surface. These long CDR H3s allow cows to mount a particularly robust immune response when immunized with viral antigens, particularly to broadly neutralizing epitopes on a stabilized HIV gp140 trimer, which has been a challenge for other species. The unusual genetics and structural biology of cows provide for a unique paradigm for creation of immune diversity and could enable generation of antibodies against especially challenging targets and epitopes.
Keywords: Antibody; Antibody diversity; Antibody repertoire; Bovine immunoglobulin; Cow antibody; Knob; Long complementarity-determining region; Ruminant antibody; Stalk; Ultralong CDR3.
© 2018 Elsevier Inc. All rights reserved.
Figures

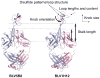

Similar articles
-
Diversity in the Cow Ultralong CDR H3 Antibody Repertoire.Front Immunol. 2018 Jun 4;9:1262. doi: 10.3389/fimmu.2018.01262. eCollection 2018. Front Immunol. 2018. PMID: 29915599 Free PMC article. Review.
-
A Broad Role for Cysteines in Bovine Antibody Diversity.Immunohorizons. 2019 Oct 16;3(10):478-487. doi: 10.4049/immunohorizons.1900058. Immunohorizons. 2019. PMID: 31619454 Free PMC article.
-
Reshaping antibody diversity.Cell. 2013 Jun 6;153(6):1379-93. doi: 10.1016/j.cell.2013.04.049. Cell. 2013. PMID: 23746848 Free PMC article.
-
Immunogenetic factors driving formation of ultralong VH CDR3 in Bos taurus antibodies.Cell Mol Immunol. 2019 Jan;16(1):53-64. doi: 10.1038/cmi.2017.117. Epub 2017 Dec 4. Cell Mol Immunol. 2019. PMID: 29200193 Free PMC article.
-
Bos taurus ultralong CDR H3 antibodies.Curr Opin Struct Biol. 2016 Jun;38:62-7. doi: 10.1016/j.sbi.2016.05.004. Epub 2016 Jun 10. Curr Opin Struct Biol. 2016. PMID: 27295423 Free PMC article. Review.
Cited by
-
Evolution of immunogenetic components encoding ultralong CDR H3.Immunogenetics. 2023 Aug;75(4):323-339. doi: 10.1007/s00251-023-01305-9. Epub 2023 Apr 21. Immunogenetics. 2023. PMID: 37084012 Free PMC article.
-
Estimates of Sequences with Ultralong and Short CDR3s in the Bovine IgM B Cell Receptor Repertoire Using the Long-read Oxford Nanopore MinION Platform.Immunohorizons. 2024 Sep 1;8(9):635-651. doi: 10.4049/immunohorizons.2400050. Immunohorizons. 2024. PMID: 39248806 Free PMC article.
-
Broadly Neutralizing Bovine Antibodies: Highly Effective New Tools against Evasive Pathogens?Viruses. 2020 Apr 22;12(4):473. doi: 10.3390/v12040473. Viruses. 2020. PMID: 32331321 Free PMC article. Review.
-
Molecular Dissection of the Antibody Response: Opportunities and Needs for Application in Cattle.Front Immunol. 2020 Jun 12;11:1175. doi: 10.3389/fimmu.2020.01175. eCollection 2020. Front Immunol. 2020. PMID: 32595642 Free PMC article. Review.
-
Immunoglobulin gene loci structure and diversity of raccoon dog (Nyctereutes procyonoides).BMC Genomics. 2025 Apr 29;26(1):424. doi: 10.1186/s12864-025-11574-1. BMC Genomics. 2025. PMID: 40301716 Free PMC article.
References
-
- Al-Lazikani B, Lesk AM, Chothia C. Standard conformations for the canonical structures of immunoglobulins. J Mol Biol. 1997;273:927. - PubMed
-
- Anderson D, Billingham RE, Lampkin GH, Medawar PB. The use of skin grafting to distinguish between monozygotic and dizygotic twins in cattle. Heredity. 1951;5:379.
-
- Arun SS, Breuer W, Hermanns W. Immunohistochemical examination of light-chain expression (lambda/kappa ratio) in canine, feline, equine, bovine and porcine plasma cells. Zentralbl Veterinarmed A. 1996;43:573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

