Pericyte ALK5/TIMP3 Axis Contributes to Endothelial Morphogenesis in the Developing Brain
- PMID: 29456135
- PMCID: PMC5871595
- DOI: 10.1016/j.devcel.2018.01.018
Pericyte ALK5/TIMP3 Axis Contributes to Endothelial Morphogenesis in the Developing Brain
Erratum in
-
Pericyte ALK5/TIMP3 Axis Contributes to Endothelial Morphogenesis in the Developing Brain.Dev Cell. 2018 Nov 5;47(3):388-389. doi: 10.1016/j.devcel.2018.10.019. Dev Cell. 2018. PMID: 30399337 Free PMC article. No abstract available.
Abstract
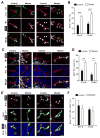
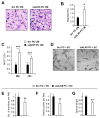
The murine embryonic blood-brain barrier (BBB) consists of endothelial cells (ECs), pericytes (PCs), and basement membrane. Although PCs are critical for inducing vascular stability, signaling pathways in PCs that regulate EC morphogenesis during BBB development remain unexplored. Herein, we find that murine embryos lacking the transforming growth factor β (TGF-β) receptor activin receptor-like kinase 5 (Alk5) in brain PCs (mutants) develop gross germinal matrix hemorrhage-intraventricular hemorrhage (GMH-IVH). The germinal matrix (GM) is a highly vascularized structure rich in neuronal and glial precursors. We show that GM microvessels of mutants display abnormal dilation, reduced PC coverage, EC hyperproliferation, reduced basement membrane collagen, and enhanced perivascular matrix metalloproteinase activity. Furthermore, ALK5-depleted PCs downregulate tissue inhibitor of matrix metalloproteinase 3 (TIMP3), and TIMP3 administration to mutants improves endothelial morphogenesis and attenuates GMH-IVH. Overall, our findings reveal a key role for PC ALK5 in regulating brain endothelial morphogenesis and a substantial therapeutic potential for TIMP3 during GMH-IVH.
Keywords: ALK5; TIMP3; blood vessel; blood-brain barrier; endothelial cells; germinal matrix hemorrhage; intracranial hemorrhage; pericytes; vascular development.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures





References
-
- Armulik A, Genove G, Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell. 2011;21(2):193–215. - PubMed
-
- Armulik A, Genove G, Mae M, Nisancioglu MH, Wallgard E, Niaudet C, He L, Norlin J, Lindblom P, Strittmatter K, Johansson BR, Betsholtz C. Pericytes regulate the blood-brain barrier. Nature. 2010;468(7323):557–61. - PubMed
-
- Arnold TD, Niaudet C, Pang MF, Siegenthaler J, Gaengel K, Jung B, Ferrero GM, Mukouyama YS, Fuxe J, Akhurst R, Betsholtz C, Sheppard D, Reichardt LF. Excessive vascular sprouting underlies cerebral hemorrhage in mice lacking alphaVbeta8-TGFbeta signaling in the brain. Development. 2014;141(23):4489–99. - PMC - PubMed
-
- Ballabh P, Xu H, Hu F, Braun A, Smith K, Rivera A, Lou N, Ungvari Z, Goldman SA, Csiszar A, Nedergaard M. Angiogenic inhibition reduces germinal matrix hemorrhage. Nat Med. 2007;13(4):477–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

