A common variant alters SCN5A-miR-24 interaction and associates with heart failure mortality
- PMID: 29457789
- PMCID: PMC5824852
- DOI: 10.1172/JCI95710
A common variant alters SCN5A-miR-24 interaction and associates with heart failure mortality
Abstract
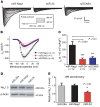
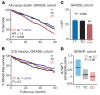
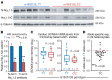
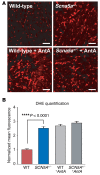
SCN5A encodes the voltage-gated Na+ channel NaV1.5 that is responsible for depolarization of the cardiac action potential and rapid intercellular conduction. Mutations disrupting the SCN5A coding sequence cause inherited arrhythmias and cardiomyopathy, and single-nucleotide polymorphisms (SNPs) linked to SCN5A splicing, localization, and function associate with heart failure-related sudden cardiac death. However, the clinical relevance of SNPs that modulate SCN5A expression levels remains understudied. We recently generated a transcriptome-wide map of microRNA (miR) binding sites in human heart, evaluated their overlap with common SNPs, and identified a synonymous SNP (rs1805126) adjacent to a miR-24 site within the SCN5A coding sequence. This SNP was previously shown to reproducibly associate with cardiac electrophysiological parameters, but was not considered to be causal. Here, we show that miR-24 potently suppresses SCN5A expression and that rs1805126 modulates this regulation. We found that the rs1805126 minor allele associates with decreased cardiac SCN5A expression and that heart failure subjects homozygous for the minor allele have decreased ejection fraction and increased mortality, but not increased ventricular tachyarrhythmias. In mice, we identified a potential basis for this in discovering that decreased Scn5a expression leads to accumulation of myocardial reactive oxygen species. Together, these data reiterate the importance of considering the mechanistic significance of synonymous SNPs as they relate to miRs and disease, and highlight a surprising link between SCN5A expression and nonarrhythmic death in heart failure.
Keywords: Cardiology; Genetic variation; Genetics; Ion channels; Noncoding RNAs.
Conflict of interest statement
Figures

Comment in
-
SCN5A: the greatest HITS collection.J Clin Invest. 2018 Mar 1;128(3):913-915. doi: 10.1172/JCI99927. Epub 2018 Feb 19. J Clin Invest. 2018. PMID: 29457788 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

