The antibiotic resistome and microbiota landscape of refugees from Syria, Iraq and Afghanistan in Germany
- PMID: 29458422
- PMCID: PMC5819293
- DOI: 10.1186/s40168-018-0414-7
The antibiotic resistome and microbiota landscape of refugees from Syria, Iraq and Afghanistan in Germany
Abstract
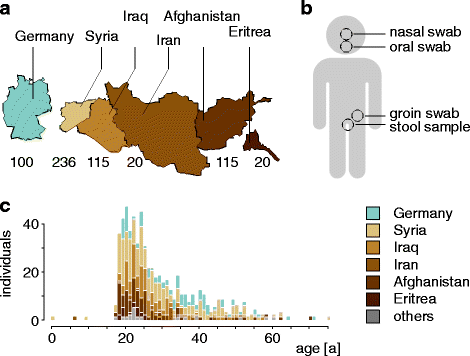
Background: Multidrug-resistant bacteria represent a substantial global burden for human health, potentially fuelled by migration waves: in 2015, 476,649 refugees applied for asylum in Germany mostly as a result of the Syrian crisis. In Arabic countries, multiresistant bacteria cause significant problems for healthcare systems. Currently, no data exist describing antibiotic resistances in healthy refugees. Here, we assess the microbial landscape and presence of antibiotic resistance genes (ARGs) in refugees and German controls. To achieve this, a systematic study was conducted in 500 consecutive refugees, mainly from Syria, Iraq, and Afghanistan and 100 German controls. Stool samples were subjected to PCR-based quantification of 42 most relevant ARGs, 16S ribosomal RNA gene sequencing-based microbiota analysis, and culture-based validation of multidrug-resistant microorganisms.
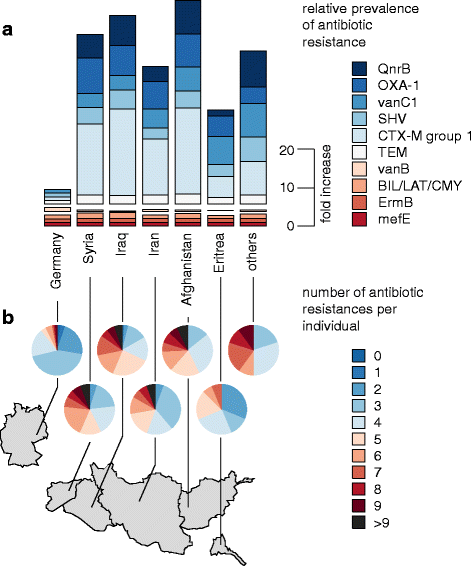
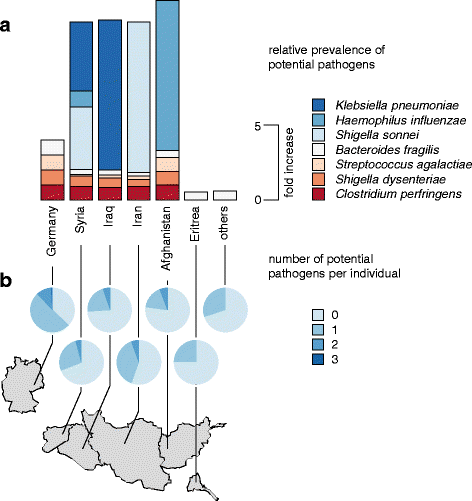
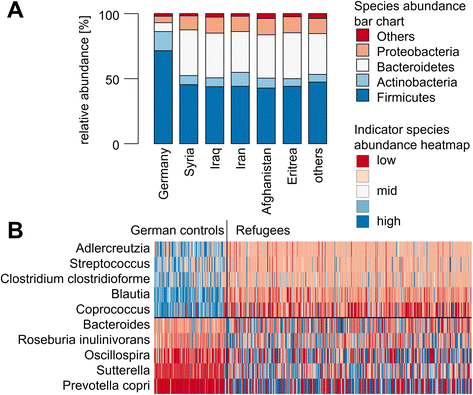
Results: The fecal microbiota of refugees is substantially different from that of resident Germans. Three categories of resistance profiles were found: (i) ARGs independent of geographic origin of individuals comprising BIL/LAT/CMA, ErmB, and mefE; (ii) vanB with a high prevalence in Germany; and (iii) ARGs showing substantially increased prevalences in refugees comprising CTX-M group 1, SHV, vanC1, OXA-1, and QnrB. The majority of refugees carried five or more ARGs while the majority of German controls carried three or less ARGs, although the observed ARGs occurred independent of signatures of potential pathogens.
Conclusions: Our results, for the first time, assess antibiotic resistance genes in refugees and demonstrate a substantially increased prevalence for most resistances compared to German controls. The antibiotic resistome in refugees may thus require particular attention in the healthcare system of host countries.
Keywords: Antibiotic resistance; Human; Refugees; Resistome.
Conflict of interest statement
Ethics approval and consent to participate
All participants gave informed consent, and the study protocol was approved by the local ethical committee at the Medical Faculty of Christian Albrecht University Kiel (D537/15; D501/14). The study does not contain identifiable information of individual persons.
Competing interests
Pius Brzoska, Jon Sherlock, and Astrid Ferlinz are employees of Thermo Fisher, but the company did not influence any aspect of the study. The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Jim O’Neil. WHO review on antimicrobial resistance. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. UK, HM Government Report: AMR.ORG; 2014 Dec p. 1–20. Report No.: main report.
-
- Section UNNS. UN News—At UN, global leaders commit to act on antimicrobial resistance [Internet]. UN News Serv. Sect. 2016 [cited 2016 Sep 23]. Available from: http://www.un.org/apps/news/story.asp?NewsID=55011#.V-Ty9DWQl0y
-
- European Centre for Disease Prevention and Control, editor. The bacterial challenge, time to react: a call to narrow the gap between multidrug-resistant bacteria in the EU and the development of new antibacterial agents. Stockholm: ECDC; 2009.
-
- Antimicrobial resistance: tackling a crisis for the health and wealth of nations. 2014.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

