Denture-associated biofilm infection in three-dimensional oral mucosal tissue models
- PMID: 29458673
- PMCID: PMC5882079
- DOI: 10.1099/jmm.0.000677
Denture-associated biofilm infection in three-dimensional oral mucosal tissue models
Abstract
Purpose: In vitro analyses of virulence, pathogenicity and associated host cell responses are important components in the study of biofilm infections. The Candida-related infection, denture-associated oral candidosis, affects up to 60 % of denture wearers and manifests as inflammation of palatal tissues contacting the denture-fitting surface. Commercially available three-dimensional tissue models can be used to study infection, but their use is limited for many academic research institutions, primarily because of the substantial purchase costs. The aim of this study was to develop and evaluate the use of in vitro tissue models to assess infections by biofilms on acrylic surfaces through tissue damage and Candida albicans virulence gene expression.
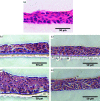
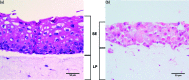
Methodology: In vitro models were compared against commercially available tissue equivalents (keratinocyte-only, SkinEthic; full-thickness, MatTek Corporation). An in vitro keratinocyte-only tissue was produced using a cancer-derived cell line, TR146, and a full-thickness model incorporating primary fibroblasts and immortalised normal oral keratinocytes was also generated. The in vitro full-thickness tissues incorporated keratinocytes and fibroblasts, and have potential for future further development and analysis.
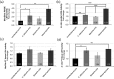
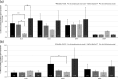
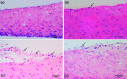
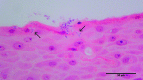
Results: Following polymicrobial infection with biofilms on acrylic surfaces, both in-house developed models were shown to provide equivalent results to the SkinEthic and MatTek models in terms of tissue damage: a significant (P<0.05) increase in LDH activity for mixed species biofilms compared to uninfected control, and no significant difference (P>0.05) in the expression of most C. albicans virulence genes when comparing tissue models of the same type.
Conclusion: Our results confirm the feasibility and suitability of using these alternative in vitro tissue models for such analyses.
Keywords: biofilm; candidosis; denture stomatitis; infection; oral mucosa; tissue model.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

