Homo- and Heterodimerization of Proteins in Cell Signaling: Inhibition and Drug Design
- PMID: 29459028
- PMCID: PMC6203451
- DOI: 10.1016/bs.apcsb.2017.08.003
Homo- and Heterodimerization of Proteins in Cell Signaling: Inhibition and Drug Design
Abstract
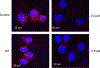
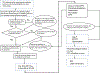
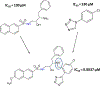
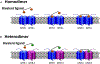
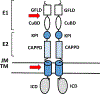
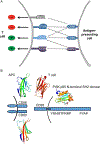
Protein dimerization controls many physiological processes in the body. Proteins form homo-, hetero-, or oligomerization in the cellular environment to regulate the cellular processes. Any deregulation of these processes may result in a disease state. Protein-protein interactions (PPIs) can be inhibited by antibodies, small molecules, or peptides, and inhibition of PPI has therapeutic value. PPI drug discovery research has steadily increased in the last decade, and a few PPI inhibitors have already reached the pharmaceutical market. Several PPI inhibitors are in clinical trials. With advancements in structural and molecular biology methods, several methods are now available to study protein homo- and heterodimerization and their inhibition by drug-like molecules. Recently developed methods to study PPI such as proximity ligation assay and enzyme-fragment complementation assay that detect the PPI in the cellular environment are described with examples. At present, the methods used to design PPI inhibitors can be classified into three major groups: (1) structure-based drug design, (2) high-throughput screening, and (3) fragment-based drug design. In this chapter, we have described some of the experimental methods to study PPIs and their inhibition. Examples of homo- and heterodimers of proteins, their structural and functional aspects, and some of the inhibitors that have clinical importance are discussed. The design of PPI inhibitors of epidermal growth factor receptor heterodimers and CD2-CD58 is discussed in detail.
Keywords: CD2–CD58; EGFR; Heterodimerization; PD-1–PD-L1; PPI inhibition; Protein–protein interaction.
© 2018 Elsevier Inc. All rights reserved.
Figures
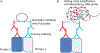

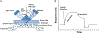
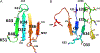
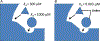


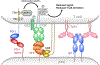
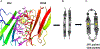
References
-
- Adams PD, Seeholzer S, & Ohh M (2002). Identification of associated proteins by coimmunoprecipitation In Protein–protein interactions: A molecular cloning manual (pp. 59–74). New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor.
-
- Aglan M, Amr K, Ismail S, Ashour A, Otaify GA, Mehrez MA, et al. (2015). Clinical and molecular characterization of seven Egyptian families with autosomal recessive robinow syndrome: Identification of four novel ROR2 gene mutations. American Journal of Medical Genetics. Part A, 167A(12), 3054–3061. 10.1002/ajmg.a.37287. - DOI - PubMed
-
- Bakail M, & Ochsenbein F (2016). Targeting protein–protein interactions, a wide open field for drug design. Comptes Rendus Chimie, 19(1), 19–27.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

