Rapid emergence of independent "chromosomal lineages" in silvered-leaf monkey triggered by Y/autosome translocation
- PMID: 29459623
- PMCID: PMC5818525
- DOI: 10.1038/s41598-018-21509-4
Rapid emergence of independent "chromosomal lineages" in silvered-leaf monkey triggered by Y/autosome translocation
Abstract
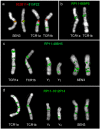
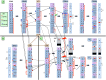
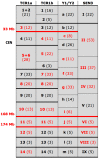
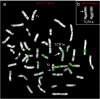
Sex/autosome translocations are rare events. The only known example in catarrhines is in the silvered-leaf monkey. Here the Y chromosome was reciprocally translocated with chromosome 1. The rearrangement produced an X1X2Y1Y2 sex chromosome system. At least three chromosomal variants of the intact chromosome 1 are known to exist. We characterized in high resolution the translocation products (Y1 and Y2) and the polymorphic forms of the intact chromosome 1 with a panel of more than 150 human BAC clones. We showed that the translocation products were extremely rearranged, in contrast to the high level of marker order conservation of the other silvered-leaf monkey chromosomes. Surprisingly, each translocation product appeared to form independent "chromosome lineages"; each having a myriad of distinct rearrangements. We reconstructed the evolutionary history of the translocation products by comparing the homologous chromosomes of two other colobine species: the African mantled guereza and the Indian langur. The results showed a massive reuse of breakpoints: only 12, out of the 40 breaks occurred in domains never reused in other rearrangements, while, strikingly, some domains were used up to four times. Such frequent breakpoint reuse if proved to be a general phenomenon has profound implications for mechanisms of chromosome evolution.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Dutrillaux B, Webb G, Muleris M, Couturier J, Butler R. Chromosome study of Presbytis cristatus: presence of a complex Y-autosome rearrangement in the male. Ann. Genet. 1984;27:148–153. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

