Blood CXCR3+ CD4 T Cells Are Enriched in Inducible Replication Competent HIV in Aviremic Antiretroviral Therapy-Treated Individuals
- PMID: 29459864
- PMCID: PMC5807378
- DOI: 10.3389/fimmu.2018.00144
Blood CXCR3+ CD4 T Cells Are Enriched in Inducible Replication Competent HIV in Aviremic Antiretroviral Therapy-Treated Individuals
Abstract
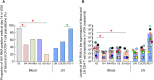
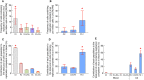
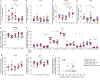
We recently demonstrated that lymph nodes (LNs) PD-1+/T follicular helper (Tfh) cells from antiretroviral therapy (ART)-treated HIV-infected individuals were enriched in cells containing replication competent virus. However, the distribution of cells containing inducible replication competent virus has been only partially elucidated in blood memory CD4 T-cell populations including the Tfh cell counterpart circulating in blood (cTfh). In this context, we have investigated the distribution of (1) total HIV-infected cells and (2) cells containing replication competent and infectious virus within various blood and LN memory CD4 T-cell populations of conventional antiretroviral therapy (cART)-treated HIV-infected individuals. In the present study, we show that blood CXCR3-expressing memory CD4 T cells are enriched in cells containing inducible replication competent virus and contributed the most to the total pool of cells containing replication competent and infectious virus in blood. Interestingly, subsequent proviral sequence analysis did not indicate virus compartmentalization between blood and LN CD4 T-cell populations, suggesting dynamic interchanges between the two compartments. We then investigated whether the composition of blood HIV reservoir may reflect the polarization of LN CD4 T cells at the time of reservoir seeding and showed that LN PD-1+ CD4 T cells of viremic untreated HIV-infected individuals expressed significantly higher levels of CXCR3 as compared to CCR4 and/or CCR6, suggesting that blood CXCR3-expressing CD4 T cells may originate from LN PD-1+ CD4 T cells. Taken together, these results indicate that blood CXCR3-expressing CD4 T cells represent the major blood compartment containing inducible replication competent virus in treated aviremic HIV-infected individuals.
Keywords: CXCR3; T follicular helper cells; circulating T follicular helper cell counterpart; lymph node; replication competent virus.
Figures




References
-
- Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A (2015) 112:E1126–34. 10.1073/pnas.1414926112 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

