Killer cell immunoglobulin-like receptor 3DL1 variation modifies HLA-B*57 protection against HIV-1
- PMID: 29461980
- PMCID: PMC5919796
- DOI: 10.1172/JCI98463
Killer cell immunoglobulin-like receptor 3DL1 variation modifies HLA-B*57 protection against HIV-1
Abstract
HLA-B*57 control of HIV involves enhanced CD8+ T cell responses against infected cells, but extensive heterogeneity exists in the level of HIV control among B*57+ individuals. Using whole-genome sequencing of untreated B*57+ HIV-1-infected controllers and noncontrollers, we identified a single variant (rs643347A/G) encoding an isoleucine-to-valine substitution at position 47 (I47V) of the inhibitory killer cell immunoglobulin-like receptor KIR3DL1 as the only significant modifier of B*57 protection. The association was replicated in an independent cohort and across multiple outcomes. The modifying effect of I47V was confined to B*57:01 and was not observed for the closely related B*57:03. Positions 2, 47, and 54 tracked one another nearly perfectly, and 2 KIR3DL1 allotypes differing only at these 3 positions showed significant differences in binding B*57:01 tetramers, whereas the protective allotype showed lower binding. Thus, variation in an immune NK cell receptor that binds B*57:01 modifies its protection. These data highlight the exquisite specificity of KIR-HLA interactions in human health and disease.
Keywords: AIDS/HIV; Innate immunity; MHC class 1; NK cells.
Conflict of interest statement
Figures
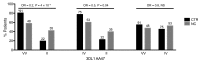

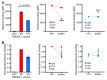
References
Publication types
MeSH terms
Substances
Grants and funding
- M01 RR000046/RR/NCRR NIH HHS/United States
- U01 AI027661/AI/NIAID NIH HHS/United States
- UM1 AI069432/AI/NIAID NIH HHS/United States
- U01 AI069470/AI/NIAID NIH HHS/United States
- P30 AI054907/AI/NIAID NIH HHS/United States
- UM1 AI069415/AI/NIAID NIH HHS/United States
- U01 AI069450/AI/NIAID NIH HHS/United States
- U01 AI069511/AI/NIAID NIH HHS/United States
- U01 AI035040/AI/NIAID NIH HHS/United States
- U01 AI069477/AI/NIAID NIH HHS/United States
- U01 AI069502/AI/NIAID NIH HHS/United States
- U01 AI069513/AI/NIAID NIH HHS/United States
- UM1 AI069494/AI/NIAID NIH HHS/United States
- UM1 AI069423/AI/NIAID NIH HHS/United States
- U01 AI035042/AI/NIAID NIH HHS/United States
- U01 AI069474/AI/NIAID NIH HHS/United States
- UL1 TR000445/TR/NCATS NIH HHS/United States
- U01 AI069434/AI/NIAID NIH HHS/United States
- U01 AI069447/AI/NIAID NIH HHS/United States
- UM1 AI069501/AI/NIAID NIH HHS/United States
- UM1 AI069472/AI/NIAID NIH HHS/United States
- U01 AI069467/AI/NIAID NIH HHS/United States
- P30 AI050409/AI/NIAID NIH HHS/United States
- U01 AI069423/AI/NIAID NIH HHS/United States
- UM1 AI069513/AI/NIAID NIH HHS/United States
- UM1 AI069424/AI/NIAID NIH HHS/United States
- R01 AI077505/AI/NIAID NIH HHS/United States
- UL1 TR001079/TR/NCATS NIH HHS/United States
- U01 AI069465/AI/NIAID NIH HHS/United States
- UM1 AI069434/AI/NIAID NIH HHS/United States
- UL1 RR024996/RR/NCRR NIH HHS/United States
- UM1 AI069495/AI/NIAID NIH HHS/United States
- U01 AI069484/AI/NIAID NIH HHS/United States
- UM1 AI069471/AI/NIAID NIH HHS/United States
- U01 AI069556/AI/NIAID NIH HHS/United States
- U01 AI038855/AI/NIAID NIH HHS/United States
- UM1 AI069428/AI/NIAID NIH HHS/United States
- UL1 RR024156/RR/NCRR NIH HHS/United States
- U01 AI069418/AI/NIAID NIH HHS/United States
- U01 AI069428/AI/NIAID NIH HHS/United States
- U01 AI069532/AI/NIAID NIH HHS/United States
- UM1 AI069439/AI/NIAID NIH HHS/United States
- P30 AI045008/AI/NIAID NIH HHS/United States
- UM1 AI069484/AI/NIAID NIH HHS/United States
- UM1 AI068634/AI/NIAID NIH HHS/United States
- U01 AI069501/AI/NIAID NIH HHS/United States
- UM1 AI069412/AI/NIAID NIH HHS/United States
- UM1 AI069470/AI/NIAID NIH HHS/United States
- U01 AI046376/AI/NIAID NIH HHS/United States
- U01 AI035041/AI/NIAID NIH HHS/United States
- R01 AI058740/AI/NIAID NIH HHS/United States
- U01 AI069432/AI/NIAID NIH HHS/United States
- UM1 AI035043/AI/NIAID NIH HHS/United States
- UM1 AI069477/AI/NIAID NIH HHS/United States
- U01 AI038858/AI/NIAID NIH HHS/United States
- U01 AI046370/AI/NIAID NIH HHS/United States
- U01 AI068636/AI/NIAID NIH HHS/United States
- P30 AI073961/AI/NIAID NIH HHS/United States
- UM1 AI069474/AI/NIAID NIH HHS/United States
- UL1 RR025777/RR/NCRR NIH HHS/United States
- U01 AI034853/AI/NIAID NIH HHS/United States
- R37 AI028568/AI/NIAID NIH HHS/United States
- UM1 AI069452/AI/NIAID NIH HHS/United States
- UM1 AI069496/AI/NIAID NIH HHS/United States
- U01 AI069415/AI/NIAID NIH HHS/United States
- UL1 TR000424/TR/NCATS NIH HHS/United States
- UL1 RR024160/RR/NCRR NIH HHS/United States
- U01 AI027658/AI/NIAID NIH HHS/United States
- U01 AI069495/AI/NIAID NIH HHS/United States
- M01 RR000051/RR/NCRR NIH HHS/United States
- U01 AI027666/AI/NIAID NIH HHS/United States
- UM1 AI069556/AI/NIAID NIH HHS/United States
- U01 AI027675/AI/NIAID NIH HHS/United States
- P30 AI110527/AI/NIAID NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- UM1 AI069502/AI/NIAID NIH HHS/United States
- U01 AI025859/AI/NIAID NIH HHS/United States
- UM1 AI069450/AI/NIAID NIH HHS/United States
- UM1 AI106701/AI/NIAID NIH HHS/United States
- UL1 RR025747/RR/NCRR NIH HHS/United States
- UM1 AI069532/AI/NIAID NIH HHS/United States
- UM1 AI069465/AI/NIAID NIH HHS/United States
- U01 AI032782/AI/NIAID NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- U01 AI069424/AI/NIAID NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- UM1 AI069419/AI/NIAID NIH HHS/United States
- R01 AI028568/AI/NIAID NIH HHS/United States
- U01 AI035039/AI/NIAID NIH HHS/United States
- U01 AI069419/AI/NIAID NIH HHS/United States
- R01 AI030914/AI/NIAID NIH HHS/United States
- UM1 AI069511/AI/NIAID NIH HHS/United States
- U01 AI069494/AI/NIAID NIH HHS/United States
- UM1 AI069447/AI/NIAID NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- U01 AI069452/AI/NIAID NIH HHS/United States
- UM1 AI069467/AI/NIAID NIH HHS/United States
- U01 AI069471/AI/NIAID NIH HHS/United States
- U01 AI069472/AI/NIAID NIH HHS/United States
- U01 AI068634/AI/NIAID NIH HHS/United States
- UM1 AI068636/AI/NIAID NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
- UM1 AI069418/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

